About NCMG
National Center for Medical Genomics
Medical genomics is a dynamically evolving scientific discipline that gathers and uses the genomic information of patients, their genetic relatives and even the entire population to identify the genetic, genomic and molecular basis of human health and disease. NCMG is a distributed scientific infrastructure, which consists of following genomic laboratories: First and Second Faculty of Medicine and Faculty of Medicine in Pilsen, Charles University in Prague, General University Hospital in Prague, Motol University Hospital; BIOCEV - Biotechnology and Biomedicine Center of the Academy of Sciences and Charles University in Vestec; Department of Internal Medicine, Hematology and Oncology (IHOK) and Centre of molecular biology and gene therapy (CMBGT) at University Hospital Brno; Central European Institute of Technology (CEITEC) at Masaryk University at Brno; Institute of Molecular and Translational Medicine at Faculty of Medicine and Dentistry at Palacký University Olomouc; University Hospital in Pilsen.
The objective of NCMG is to provide for an operation of next generation sequencing platforms and subsequent technologies for analysis of human genomes and to allow qualified usage of these technologies in biomedical research and translational medicine in Czech Republic. Different localization and partially different specialization of individual laboratories guarantee expansion of genomics in CR, corresponding with global trends. NCMG laboratories are equipped with modern instruments and provide fundamental instrumental, methodical and experimental expertise, which is necessary for genomic sequencing, whole genome genotyping, cytogenetic analysis and analysis of genome, transcriptome and epigenome. NCMG is also equipped with corresponding computational and data storage resources and offers bioinformatics and statistics support for projects with focus on study of both complex and rare diseases and also oncological diseases.
Services
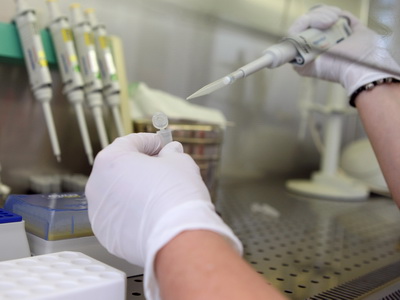
Genome sequencing and bioinformatics analysis
- NGS sequencing - genome, exome, targeted, RNA sequencing
- Microarrays - SNP and CNV detection, expression
Teaching and consulting
- We can help you with the design of your project, it's realization and with the data analysis
Access to the cohort of patients and controls
- You can download our publicly available Czech database of genomic variants
If you need any other information or want to collaborate, please contact prof. Stanislav Kmoch: skmoch@lf1.cuni.cz
NCMG database
- Download the database
Version 2
1799 samples (796 males and 1003 females)
Download the database: Excel file -- added on 26. 2. 2024
Users are obliged to acknowledge NCMG in any output (publications, patents, public presentations) that were created by using the NCMG instrumentation or expertise.
Version 1
1055 samples (442 males and 613 females)
- 572 samples, representing common czech poulation without severe diseases (neselektovana_populace)
- 483 samples, representing czech poulation older than 70 years, without severe diseases (NCMG_controls)
Download the database: Excel file -- updated on 23. 9. 2019
Users are obliged to acknowledge NCMG in any output (publications, patents, public presentations) that were created by using the NCMG instrumentation or expertise.
- File description and data processing:
variants: database contains all coding and splice variants with population frequency < 5 %
reference sequence: hg19
tools: novoalign, picard tools, GATK HaplotypeCaller, GEMINI
description of individual columns are part of the Excel file, tab "Legenda"
If you have any questions, please contact Viktor Stránecký (vstra@lf1.cuni.cz) or Anna Přistoupilová (apris@lf1.cuni.cz)
News
Added list of publications linked with NCMG for the year 2025 and updated overview of ongoing projects with the support of NCMG infrastructure - 09.01.2026
Martina Živná received the Czech Minister of Health Award for Medical Research and Development in 2025 for her successful project Identification and Characterization of Genetic Factors of Hereditary Tubulointerstitial Kidney Disease II.
The awards were presented on December 9, 2025, honoring 10 top-rated research projects supported by the Czech Health Research Council (AZV ČR) and successfully completed in 2024. The awards were presented by Minister of Health Prof. Vlastimil Válek together with the agency's chairman, Prof. Ondřej Slabý.
More details are here.

Photo: Kmoch Team (HT)
18.12.2025
Added updated list of recent publications linked with NCMG for the period 1-11/2025 - 27.11.2025
Updated overview of ongoing projects with the support of NCMG infrastructure, publications from the period of 1-6/2025 and list of available technologies - 30.06.2025
Added updated list of 2024 publications and recent publications for the period 1-4/2025 - 10.04.2025
Updated list of current projects - 28.1.2024
Added list of publications supported by NCMG for the year 2024 - 13.12.2024
We have updated the database of genomic variants. Now containing 1799 samples, of which 1003 are women and 796 are men. All coding variants and variants affecting splicing are included. - 26/02/2024
Added updated list of recent publications linked with NCMG for the year 2023 - 05.01.2024
New dedication for the next period: "The National Center for Medical Genomics" or "The NCMG research infrastructure" (LM2023067) added. - 30.10.2023
Added updated list of recent publications and projects linked with NCMG for 2023 - 11.8.2023
Updated list of projects linked with NCMG - 14.02.2023
New publications supported by NCMG added - 16.01.2023
New publications supported by NCMG added - 22. 11. 2021
NCMG and COVID-19 testing - 9. 10. 2020
Stanislav Kmoch on know-how that made testing for Covid safer
Professor Stanislav Kmoch has been researching rare diseases for thirty years. In the current coronavirus pandemic, knowledge from his laboratory, which is part of the NCMG, has significantly contributed to the development of a new procedure for the diagnosis of covid-19 and other viral diseases. "It's a secondary but nice result of our many years of research into rare diseases," Kmoch describes the creation of the university spin-off GeneSpector.
GeneSpector provides a unique solution for rapid Covid-19 detection.
- viRNAtrap™ Collection Tube inactivates the infectious agents developed for save operations at ambient temperature
- viRNAtrap™ Extraction Kit is a best-in-class solution for fast and high throughput extraction at any open automated platform or when using manual work-up
- high-quality PCR kits for fast COVID-19 detection
- triplex COVID-INFA-INFB (to be launched in October 2020)
New publications supported by NCMG added - 22. 10. 2020
Since 2020, NCMG has a new project number: LM2018132. Please, use in your publications this number. - 28. 5. 2020
New publications supported by NCMG added - 28. 5. 2020
NCMG and COVID-19 testing - 14. 4. 2020
A wide academic community has joined an initiative that aims to help with laboratory testing for COVID-19. Researchers from several departments of the 1st Medical Faculty of Charles University - from the Department of Pediatrics and Adolescent Medicine, Department of Biochemistry and Experimental Oncology, Department of Biology and Medical Genetics and Department of Immunology and Microbiology and the National Center of Medical Genomics developed in 14 days functional system for the determination of covid19 in clinical material. This system has been used by the VFN for two weeks to test employees and admitted patients. As stated by prof. Ing. Stanislav Kmoch, CSc., Currently about 100 samples are examined daily. The much higher capacity of the established procedure is so far limited by the logistics of sampling, administration and the demands on the processing of potentially infectious material. Employees of the 1st Faculty of Medicine of Charles University thus help to increase the capacity in testing for COVID-19 in the Czech Republic and together with VFN significantly contribute to managing epidemiological problems in this difficult time, adds another scientist, prof. RNDr. Libuše Kolářová, CSc.
- IMTM declares war against COVID-19
- CEITEC Scientists Are Developing Faster Tests for COVID-19 Disease
New publications supported by NCMG added - 6. 1. 2020
New publications supported by NCMG added - 5. 11. 2019
We published a new version of Czech database of genomic variants - it contains 1055 samples (442 males a 613 females) - 23. 9. 2019
New publications supported by NCMG added - 23. 9. 2019
New publication supported by NCMG added - 19. 6. 2019
New publication supported by NCMG added - 11. 6. 2019
New publications supported by NCMG added - 4. 6. 2019
We upgraded our NGS sequencing workflow by installing a new high throughput sequencer - Illumina NovaSeq 6000 - 4. 3. 2019
New publications supported by NCMG added - 5. 12. 2018
New publications supported by NCMG added - 20. 9. 2018
New publications supported by NCMG added - 12. 7. 2018
New version of our database of genomics variants - containing 468 samples from common population without severe diseases (252 males and 216 females) - 27. 4. 2018
New publications supported by NCMG added - 25. 4. 2018
We would like to invite you to attend and participate in the 2nd HVP Variant Effect Prediction Training Course that will be held from Monday 6 to Wednesday 8 November in the beautiful historic city of Prague, Czech Republic. More information and registration at http://vep.variome.org/. You can still register for a discounted price - Earlybird ends 11.59 PM CET 31st August 2017!

New publications supported by NCMG added - 28. 8. 2017
Two teams from NCMG have received Minister of Health of Czech Republic Awards for research and development in medicine for 2016. Team of doc. Ing. Stanislav Kmoch, CSc. was awarded for the project " Identification of the genetic and molecular basis of rare genetic disorders using novel genomic methods" and the team of prof. RNDr. Šárka Pospíšilová, Ph.D. for the project "Molecular characterization of B-cell receptors and its relation to the evolution of genetic changes in chronic lymphocytic leukemia." - 16. 12. 2016
We would like to introduce you our Scientific Advisory Board - 2. 12. 2016
Please visit our new social networks profiles on Facebook and Twitter. - 4. 10. 2016
Key people
Prof. Ing. Stanislav Kmoch, CSc.
For First Faculty of Medicine, Charles University in Prague and General University Hospital in Prague; head of the research program Development of Diagnostic and Therapeutic Procedures at BIOCEV and head of Research Unit for Rare Diseases, First Faculty of Medicine and General University Hospital. His main focus lies in the study of genetic, molecular, cellular and pathologic causes of rare and complex diseases.
Prof. MUDr. Milan Macek, DrSc., MHA

For Second Faculty of Medicine, Charles University in Prague and University Hospital Motol; chairman of the largest academic medical/molecular genetics institution in the Czech Republic - Department of Biology and Medical Genetics; ÚBLG. He is also the past President of the European Society of Human Genetics (ESHG.org; 2011), past-board member of the European Cystic Fibrosis Society (ECFS.eu; 2007-2015 and of the European Society for Human Reproduction and Embryology (ESHRE.eu; 2012-2014). He also serves at the Commission Expert Group on Rare Diseases (formerly EUCERD.eu). ÚBLG was designated by the Czech Ministry of Health as a National Coordination centre for rare diseases (Bulletin 4/2012) which serves as a "clearing centre" for the dissemination of knowledge in rare disease-related genomics gathered within various international / European projects such as CF Thematic Network, EuroGentest, EuroCareCF, Techgene.eu, RD-connect.eu, Norway Grants or EU ESF funds. His main research and clinical interest is genomics of rare diseases, and how to translate such knowledge to clinical diagnostics and/or bedside. Prof. Macek is also the Czech National coordinator of Orpha.net and member of the Diagnostic Committee of the International Rare Disease Consortium (IRDIRC.org).
Prof. RNDr. Šárka Pospíšilová, Ph.D.

For Masaryk University and University Hospital Brno; research groupleader of Medical Genomics, coordinator of Molecular Medicine program at CEITEC, and head of Centre of molecular biology and gene therapy of University Hospital Brno. Research projects focus primarily on study of p53 signalling pathway and other molecular markers of hemato-oncological diseases, mutational and functional analyses of immunoglobulin genes and microRNAs in lymphoproliferative disorders. The laboratory is also intensively on application of genomic approaches in diagnostics of hematologic and oncologic diseases (leukemias, lymphomas, breast cancer and other malignancies) in postnatal but also in prenatal and pre-implantation diagnostics.
prim. MUDr. Ivan Šubrt, Ph.D.

For the Faculty of Medicine in Pilsen, Charles University Prague and the University Hospital in Pilsen; head of the Department of Medical Genetics and of the research group Medical Genomics. Research projects are focused mainly on the study of molecular markers of disorders of human reproduction, susceptibility to infectious diseases, oncogenesis and on the utilization of genomic technologiesfor diagnosis of rare genetic diseases.
Assoc. Prof. Marian Hajduch, MD, Ph.D.

Faculty of Medicine and Dentistry, Palacky University Olomouc and University Hospital Olomouc. Director of the Institute of Molecular and Translational Medicine, head of the research program Chemical biology and experimental therapeutics. Research projects are focused on research into the molecular biology of cancer in particular, on research and development of new drugs and diagnostic methods and translational medicine.
Institutions

Research Unit for Rare Diseases, First Faculty of Medicine at Charles University in Prague, General University Hospital and BIOCEV
The laboratory is focused on studying the molecular basis of rare genetically determined disorders. In its work combines latest techniques and methods of medical genomics, bioinformatics, metabolomics, molecular biology and biochemistry with detailed clinic-pathologic evaluation of the studied diseases. In recent years this approach led to identification of causal genes and characterization of molecular basis of more than a dozen of severe genetic diseases. The laboratory offers it's expertise to clinical centers both in the Czech Republic and abroad.

Department of Biology and Medical Genetics, Second Faculty of Medicine, Charles University and University Hospital Motol
Department of Biology and Medical Genetics is a clinical-diagnostic, academic and research institution. Main topics are diagnostics and further study of hereditary diseases in prenatal and postnatal phase of development by means of state of the art techniques of molecular genetics, molecular cytogenetics and next generation sequencing. By decision of Czech Ministry of Health a National coordination center for rare diseases was established at Department of Biology and Medical Genetics.

Masaryk University - CEITEC
CEITEC MU was established as a part of scientific centre of excellence Central European Institute of Technology (CEITEC). Shared Core Facility Genomics and Laboratory of Medical Genomics are based in newly built premises University Campus that offers high-end equipment and optimal conditions to solve current issues of biomedical and clinical research (from genome analysis, mapping of key genetic defects, molecular genetic diagnostics to search for suitable targets for gene therapy). The portfolio of services of shared Genomics laboratory includes high throughput massively parallel sequencing, cell separation using FACS and MACS, real time PCR including microfluidic PCR, microarrays and other technologies. Scientists from the Czech and foreign universities and research institutes interested in using the services of Genomics Laboratory can apply for financial support from the Ministry of Education, Youth and Sports (MEYS).

University Hospital Brno
Centre of molecular biology and gene therapy (CMBGT) forms a part of Department of Internal Medicine, Hematology and Oncology (IHOK) focused on research and development and is one of the key units of the University Hospital Brno. It deals with the molecular biological diagnostics in the field of haematooncology, solid tumors, inherited diseases and identification of pathogens in immunocompromised patients. It also applies cytometry and cytogenetic diagnostics in hematology and hematooncology an performs applied research in general field of human medicine.

Department of Medical Genetics, Faculty of Medicine in Pilsen, Charles University Prague and the University Hospital in Pilsen
Research activities of the group focus on the study of molecular markers of disorders of human reproduction, susceptibility to infectious diseases, oncogenesis and on the clinical aspects of broad group of rare genetic diseases.

Institute of Molecular and Translational Medicine,
Faculty of Medicine and Dentistry, Palacký University in Olomouc
The Institute of Molecular and Translational Medicine, Faculty of Medicine and Dentistry, Palacký University in Olomouc is cutting-edge biomedical research institute in the Czech Republic. The IMTM’s mission is basic and translational biomedical research with the goal to understand the underlying causes of cancer and infectious diseases and to develop future human medicines and diagnostics.
Research teams
Research Unit for Rare Diseases, First Faculty of Medicine at Charles University in Prague, General University Hospital and BIOCEV
Head of the group:
Prof. Ing. Stanislav Kmoch, CSc.
stanislav.kmoch@lf1.cuni.cz
+420 22496 7016, +420 22496 7691
NGS sequencing:
- RNDr. Hana Hartmannová, Ph.D., hana.hartmannova@lf1.cuni.cz; main coordinator
- Ing. Kateřina Hodaňová, Ph.D., katerina.hodanova@lf1.cuni.cz; coordinator
- Mgr. Barbora Bendová, barbora.bendova@natur.cuni.cz; PhD. coordination
- Ing. Lenka Steiner Mrázová, Ph.D., lenka.mrazova@lf1.cuni.cz; RNA sequencing
- RNDr. Alena Vrbacká, Ph.D., alena.cizkova@lf1.cuni.cz; long-read sequencing
- Ing. Lenka Piherová, PhD., lenka.piherova@lf1.cuni.cz
Bioinformatics, biostatistics:
- Mgr. Václav Janoušek, PhD., vaclav.janousek@lf1.cuni.cz
- Ing. Dita Mušálková, PhD., Dita.Musalkova@lf1.cuni.cz
- Ing. Petr Nehasil, petr.nehasil@lf1.cuni.cz
- Mgr. Viktor Stránecký, Ph.D., viktor.stranecky@lf1.cuni.cz
- Mgr. et Mgr. Anna Přistoupilová, Ph.D., anna.pristoupilova@lf1.cuni.cz
- Mgr. Petra Zemánková (Boudová), Ph.D., petra.boudova@lf1.cuni.cz
Laboratory technician:
- Helena Trešlová, Helena.Myskova@lf1.cuni.cz
- Miroslav Votruba, miroslav.votruba@lf1.cuni.cz
Technical and administrative support:
-
Ing. Eva Oliveriusová, estar@lf1.cuni.cz, +420 22496 7680
Ethics:
-
Mgr. Věra Franková, Ph.D., vera.frankova@lf1.cuni.cz, +420 22496 7224
Department of Biology and Medical Genetics, Second Faculty of Medicine, Charles University and University Hospital Motol
Head of the research team:
Prof. MUDr. Milan Macek, DrSc., MHA
milan.macek.jr@lfmotol.cuni.cz
+420 224 433 500
Deputy head of the research team:
Prof. Ing. Zdeněk Sedláček, DrSc.
zdenek.sedlacek@lfmotol.cuni.cz
+420 257 296 153
Molecular genetics, next generation sequencing:
- RNDr. Petra Peldová, Ph.D., petra.peldova@fnmotol.cz, +420 2 2443 3529
- Mgr. Malgorzata Libik, Ph.D., Malgorzata.libik@fnmotol.cz, +420 2 2443 3545
- Mgr. Markéta Wayhelová, Ph.D., marketa.wayhelova@fnmotol.cz, +420 2 2443 3517
- Doc. MUDr. Alice Krebsová, Ph.D., krea@ikem.cz,
- Mgr. Miroslava Hančárová, PhD., miroslava.hancarova@lfmotol.cuni.cz, +420 257 29 6152
- Mgr. Pavel Votýpka - pavel.votypka@fnmotol.cz, +420 22443 3594
- Mgr. Lenka Dvořáková - lenka.dvorakova@fnmotol.cz, +420 22443 3545
- Ing. Hana Řezáčová - hana.zunova@fnmotol.cz, +420 22443 3568
- Mgr. Zuzana Slámová - zuzana.slamova@fnmotol.cz, +420 22443 3562
- Priv. Doz. MUDr. Dana Thomasová, Ph.D. - dana.thomasova@fnmotol.cz, +420 22443 5992
Bioinformatics:
- Mgr. Jan Geryk, Ph.D., jan.geryk@fnmotol.cz, +420 224 433 528
- MUDr. Marek Turnovec, marek.turnovec@lfmotol.cuni.cz, +420 224 433 535
Molecular Medicine Program CEITEC MU – Genomics Core Facility, Central Laboratory Bioinformatics and Research Group of Medical Genomics
Head Research Group of Medical Genomics:
prof. RNDr. Sarka Pospisilova, Ph.D.
sarka.pospisilova@ceitec.muni.cz
+420 549497917
Head of Genomics Core Facility:
MVDr. Boris Tichy, Ph.D.
boris.tichy@ceitec.muni.cz
+420 733141527
Head of CF Bioinformatics:
Mgr. Vojtěch Bystrý, Ph.D.
vojtech.bystry@ceitec.muni.cz
+420 549493651
NGS, single cells microarrays:
- MVDr. Boris Tichý, Ph.D., boris.tichy@ceitec.muni.cz, +420 733141527
- Mgr. Filip Pardy, filip.pardy@ceitec.muni.cz, +420 549498317
- Mgr. Terézia Kurucová, terezia.kurucova@ceitec.muni.cz, +420549498196
Biostatistics, bioinformatics:
- Mgr. Vojtěch Bystrý, Ph.D., vojtech.bystry@ceitec.muni.cz, +420 549493651
- Nicolas Blavet, Ph.D., nicolas.blavet@ceitec.muni.cz, +420 549498964
- Mgr. Martin Demko, Ph.D., martin.demko@ceitec.muni.cz, +420 549498964
Contact e-mail addresses:
- CF Genomika: cfg@ceitec.muni.cz
- CF Bioinformatika: vojtech.bystry@ceitec.muni.cz
- CF administrative support: core.facility@ceitec.muni.cz
Websites of CF Genomics:
CEITEC MU: cfg.ceitec.cz
Bioinformatics: https://www.ceitec.eu/bioinformatics-core-facility/cf284
Centre of Molecular Biology and Gene Therapy of University Hospital Brno (CMBGT)
Head of CMBGT:
prof. RNDr. Sarka Pospisilova, Ph.D.
sarka.pospisilova@fnbrno.cz
+420 532234622
Unit of congenital and genetic diseases:
- doc. RNDr. Lenka Fajkusová, CSc., lfajkusova@fnbrno.cz, +420 532234625
Unit of Cancer Genomics:
- RNDr. Jitka Malčíková, Ph.D., jmalcikova@fnbrno.cz, +420 532234396
- Mgr. Karla Plevová, Ph.D., kplevova@fnbrno.cz, +420 532234396
Department of Medical Genetics, Faculty of Medicine in Pilsen, Charles University Prague and the University Hospital in Pilsen
Head of the research team:
MUDr. Ivan Šubrt, Ph.D.
subrti@fnplzen.cz
+420 377 402 800
Molecular biology, genetics, NGS sequencing and data analysis:
- MUDr. Monika Černá, Ph.D., cernam@fnplzen.cz, +420 377 402 810
- Mgr. Markéta Hejnalová, hejnalovam@fnplzen.cz, +420 377 402 810
- Ing. Pavla Komrsková, Ph.D., komrskovap@fnplzen.cz, +420 377 402 810
- Mgr. Lukáš Strych, strychl@fnplzen.cz, +420 377 402 810
- Mgr. Tomáš Zavoral, zavoralt@fnplzen.cz, +420 377 402 810
- Mgr. Nikol Jedličková
- Ing. Jitka Tejcová
Multiomics, biomarkers:
- Mgr. Viktor Hlaváč, Ph.D., viktor.hlavac@lfp.cuni.cz, +420 377 593 840
Clinical research, cytogenomics:
- MUDr. Radka. Jaklová, jaklova@fnplzen.cz, +420 377 402 870
- RNDr. Petra. Vohradská, vohradskap@fnplzen.cz, +420 377 402 803
- Ing. Kateřina Tesařová, tesarovak@fnplzen.cz, +420 377 402 803
Institute of Molecular and Translational Medicine
Leader of the research team:
Assoc. Prof. Marian Hajduch, MD, PhD
marian.hajduch@upol.cz
+420 585632082, +420 585632083
Molecular biology, genetics, NGS sequencing:
- Josef Srovnal, MD, PhD, josef.srovnal@upol.cz, +420 585632137
- Assoc. Prof. Jiri Drabek, PhD, jiri.drabek@upol.cz, +420 585632070
- Radek Trojanec, PhD, radek.trojanec@upol.cz, +420 585632089
Bioinformatics, biostatistics:
- Mgr. Jana Vrbková, Ph.D., jana.vrbkova@upol.cz, +420 585632057
Our projects
Research Unit for Rare Diseases, First Faculty of Medicine at Charles University in Prague, General University Hospital and BIOCEV
NW26-07-00090 Identification and characterization of genetics facotrs of hereditary tubulointerstitial kidney disease III; Grant provider: Grant provider: Ministry of Health of the Czech Republic; Main recipient: Charles University – First Faculty of Medicine; Project lead: Mgr. Martina Živná, Ph.D.; Period of research project: 2026-2029
26-21355S Heterodimerization of human GLP-G9a histone methyltransferase and abnormal epigenetics in Kleefstra syndrome; Grant provider: The Czech Science Foundation (GACR); Main recipient: Charles University – First Faculty of Medicine; Project lead: Ing. Aleš Hnízda Ph.D.; Period of research project: 2026-2029
26-21263S Enhancing PRPF31 expression as a therapeutic strategy for retinitis pigmentosa 11; Grant provider: The Czech Science Foundation (GACR); Main recipient: Charles University – First Faculty of Medicine; Project lead: prof. David Staněk, Ph.D.; Period of research project: 2026-2029
CZ.02.01.01/00/23_020/0008540 - Multi-omics platform for the search for biological correlates of diseases and the development of new diagnostic, preventive and therapeutic procedures (MULTIOMICS.CZ); Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University - First Faculty of Medicine; Project lead: prof. Ing. Stanislav Kmoch CSc.; Period of research project: 2025 – 2028
LUAUS25093 - Genetic causes and therapy of rare hereditary amyloidoses, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University - First Faculty of Medicine; Project lead: MUDr. Jakub Sikora, Ph.D; Period of research project: 2025 – 2028
LUAUS24087 - Identification and characterization of genetic causes of hereditary tubulointerstitial kidney diseases, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University - First Faculty of Medicine; Project lead: prof. Ing. Stanislav Kmoch CSc.; Period of research project: 2024 – 2028
NW24-04-00067 Hereditary movement disorders: genetic variability of parkinsonian and hyperkinetic syndromes, Grant provider: MZ0 – Ministry of Health of the Czech Republic; Main recipient: General University Hospital in Prague (VFN) and Charles University - First Faculty of Medicine; Project leaders: Doc. MUDr. Jiří Klempíř, Ph.D and MUDr. Jakub Sikora, Ph.D; Period of research project: 2024 – 2027
UNCE - 24/MED/022 - Center for the Study of Rare Genetic Disorders II, Grant provider: Charles University (university grant UNCE – University research centers), Main recipient: First Faculty of Medicine; Project lead: prof. Ing. Stanislav Kmoch CSc.; Period of research project: 2024-2029
NW25-07-00303 - Optimization of corneal endothelial failure diagnostics in clinical practice and transplantology; Grant provider: Ministry of Health of the Czech Republic; Main recipient: Charles University - First Faculty of Medicine; Project lead: Ing. Ľubica Ďuďáková, PhD; Period of research project: 2025 – 2028
NU23-08-00123 - Improving diagnostics and treatment in patients with ultra-rare diseases using advanced forms of genomic analysis and artificial intelligence, in connection with new methods of digitizing their phenotype; Grant provider: AZV - Agency for Health Research of the Czech Republic; Main recipient: Charles University - First Faculty of Medicine; Project lead: prof. MUDr. Petra Lišková, MD, Ph.D; Period of research project: 2025 – 2028, Note: Only the project proposal was found.
GA24-10324S - Exploring the Molecular Basis of Anterior Segment Dysgenesis using Advanced Sequencing and Organoid Technology; Grant provider: Czech Science Foundation; Main recipient: Charles University - First Faculty of Medicine; Project lead: prof. MUDr. Petra Lišková, MD, Ph.D; Period of research project: 2024 - 2026
NW24-03-00092 - Variants of uncertain significance in the CHEK2 gene: their functional classification and characterization of tumors in carriers of pathogenic mutations II, Grant provider: MZ0 – Ministry of Health of the Czech Republic; Main recipient: Institute of Molecular Genetics of the Czech Academy of Sciences, General University Hospital in Prague (VFN) and Charles University - First Faculty of Medicine; Project leaders: MUDr. Libor Macůrek, PhD., MUDr. Petra Kleiblová, prof. MUDr. Zdeněk Kleibl, PhD., Period of research project: 2024 – 2027
RVO-VFN 64165 (subproject GIP-23-L-09-846) – Idenitification of Genetic Oncologic Risks (IGOR); Grant provider: Ministry of Health of the Czech Republic; Main recipient: General Faculty Hospital in Prague Charles University - First Faculty of Medicine; Institutional Support – Project for the Long-Term Strategic Development of a Research Organization; Project lead: prof. MUDr. Zdeněk Kleibl, PhD.
SVV260631 - Methodology of Biomedical Research (Student Scientific Project); Grant provider: Charles University; Main recipient: First Faculty of Medicine; Project lead: prof. Ing. Stanislav Kmoch CSc.;
Collaboration between the First Faculty of Medicine, Charles University, and the Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences on the project for the development of a repository for sharing genomic and medical data within the European FEGA platform. (18.12.2024)
NU23-07-00281 Characterization of the molecular basis of rare genetic diseases of pediatric onset using new methods of genome analysis II. Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: prof. Ing. Stanislav Kmoch, CSc.; Period of research project: 2023 - 2026
LM2023067 The National Center for Medical Genomic, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University in Prague / First Faculty of Medicine, Recipient: prof. Ing. Stanislav Kmoch, CSc. Period of research project: 2023 - 2026
TN02000132 National centre for new methods of diagnosis, monitoring, treatment and prevention of genetic diseases, Grant provider: Technology Agency of the Czech Republic Main recipient: Institute of Molecular Genetics of the Czech Academy of Sciences, Recipient: PD. Dr. rer. nat. ha Radislav Sedláček, Period of research project: 2023 - 2026
NU23-01-00500 Identification and characterization of genetic factors and pathogenetic mechanism of defects affecting de novo purine metabolism, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: Ing. Marie Zikánová, PhD.; Period of research project: 2023 – 2026
NU23-03-00150 Classification of variants of unknown significance in cancer susceptibility genes by the multiplex PCR/NGS-based analysis of their impact on pre-mRNA splicing (II)), Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: General University Hospital in Prague (VFN) ; Recipient : MUDr. Petra Kleiblová, Ph.D., Period of research project: 2023 – 2026
GA25-16979S - Spatially resolved single-cell transcriptomic map of human ischemic brain; Grant provider: Czech Science Foundation; Main recipient: Institute of Biotechnology of the Czech Academy of Sciences; Project lead: Ing. Lukáš Valihrach Ph.D.; Period of research project: 2025 – 2027
GA24-11364S - A humanized organoid-based platform for stroke modelling and mRNA-based therapeutic intervention; Grant provider: Czech Science Foundation; Main recipient: Institute of Biotechnology of the Czech Academy of Sciences; Project lead: Ing. Lukáš Valihrach Ph.D.; Period of research project: 2024 – 2026
10112569 - Activation and switch of fates in T lymphocytes; Grant provider: Horizon Europe; Main recipient: Institute of Molecular Genetics of the Czech Academy of Sciences; Project lead: Mgr. Ondřej Štěpánek, Ph.D.; Period of research: 2025 - 2029
Strategie AV21 – Fungi - new threats and opportunities; Grant provider: Czech Academy of Sciences; Coordinating Centre: Institute of Microbiology of the Czech Academy of Sciences; Project coordinator: Mgr. Miroslav Kolařík, Ph.D.; Period of research project: 2024 - 2028
Solved projects
NU22-03-00276 - Functional classification of the TP53 gene variants with distinct penetrance and its use for better management of cancer prevention in mutation carriers, Grant provider: MZ0 – Ministry of Health of the Czech Republic; Main recipient: Institute of Molecular Genetics of the Czech Academy of Sciences, General University Hospital in Prague (VFN), Project leaders: MUDr. Libor Macůrek, PhD., MUDr. Petra Kleiblová; Period of research project: 2022-2025
LX22NPO5107 National Institute for Neurological Research, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: St. Anne's University Hospital in Brno, Recipient: prof. MUDr. Milan Brázdil, Ph.D., Period of research project: 2022-2025
LX22NPO5104 National Institute for Research of Metabolic and Cardiovascular Diseases, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Institute for Clinical and Experimental Medicine, Recipient: : prof. MUDr. Martin Haluzík, DrSc., Period of research project: 2022 – 2025
LX22NPO5102 National institute for cancer research, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University in Prague / First Faculty of Medicine, Recipient: prof. MUDr. Aleksi Šedo, Period of research project: 2022 – 2025
NU22-07-00474 Molecular genetic causes and biochemical consequences of Congenital disorders of glycosylation, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: RNDr. Hana Hansíková, CSc., Period of research project: 2022 – 2025
GA23-05327S - Astrocyte-microglia communication as a target for stroke therapy; Grant provider: Czech Science Foundation; Main recipient: Institute of Biotechnology of the Czech Academy of Sciences; Project lead: Ing. Lukáš Valihrach Ph.D.; Period of research project: 2023 – 2025
NU22-03-00182 - Circulating Tumor DNA and its Prognostic Impact in Hodgkin Lymphoma; Grant provider: Ministry of Health of the Czech Republic; Main recipient: Faculty Hospital Královské Vinohrady; Other recipient: Charles University - First Faculty of Medicine; Project leads: MUDr. Heidi Móciková Ph.D., MUDr. Ondřej Havránek Ph.D.; Období řešení projektu: 2022 – 2025
NU21-03-00411 - Early identification of prognostic factors connected with worse outcome of Non-Hodgkin’s Lymphoma; Grant provider: Ministry of Health of the Czech Republic; Main recipient: General Faculty Hospital in Prague; Other recipient: Charles University - First Faculty of Medicine; Project lead: prof. MUDr. Marek Trněný CSc.; Period of research project: 2021 – 2025
NU22-03-00210 - Genomic characterization of adult B-precursor acute lymphoblastic leukemia for prediction of treatment response to targeted therapy; Grant provider: Ministry of Health of the Czech Republic; Main recipient: Institute of Hematology and Blood Transfusion; Project lead: doc. MUDr. Cyril Šálek Ph.D.; Period of reasearch project: 2022 – 2025
NU22-07-00614 Mitochondrial optic neuropathies: a complex genetic and metabolomic analysis to characterize molecular bases and to identify novel biomarkers, Grant provider: Ministry Of Health Of The Czech Republic, Main recipient: Charles University / 1st Medicine faculty; Recipient: prof. MUDr. Petra Lišková, MD, Ph.D., Period of research project: 2022 – 2025
NU21-07-00033 Identification and characterization of genetic factors contributing to inherited tubulointerstitial kidney disease II. Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: Mgr. Martina Živná, Ph.D. Period of research project: 2021-2024
NU21-08-00324 Evaluation of new biomarkers of Fabry disease in selected groups of patients in the Czech Republic, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: RNDr. Ladislav Kuchař, Ph.D. Period of research project: 2021 – 2024
NU21-07-00225 - Spectrum of NGS detected somatic mutations and their association with prognosis and outcome of adolescent and young adult patients with Ph-positive leukemia; Grant provider: Ministry of Health of the Czech Republic; Main recipient: Institute of Hematology and Blood Transfusion; Project lead: doc. Mgr. Kateřina Machová Poláková Ph.D.; Period of research project: 2021 – 2024
NU22-A-123 Prognostic factors affecting treatment outcomes and mortality in COVID-19 patients, new diagnostic methods, current and new terapeutic opportunities for COVID-19, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Thomayer University Hospital, Recipient: Doc. MUDr. Roman Zazula, Ph.D., Period of research project: 2022 – 2023
NU20-03-00285 BIOINFORMATICS AND FUNCTIONAL ANALYSES OF SUSCEPTIBILITY VARIANTS SUPPORTING THE NGS-BASED TESTING OF HEREDITARY CANCERS IN THE CZECH REPUBLIC (II), Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: Prof. MUDr. Zdeněk Kleibl, Ph.D Period of research project: 2020 – 2023
NU20-07-00026 Changes of transcriptome during early postnatal development in humans: impact of premature birth on control of energy metabolism, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Institute of Physiology of the Czech Academy of Sciences, Recipient: : MUDr. Jan Kopecký, DrSc., Period of research project: 2020-2023
NU20-03-00283 Hereditary cancer predisposition in breast or colorectal cancer patients not indicated for routine genetic testing, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: MUDr. Petra Kleiblová, Ph.D. Period of research project: 2020 – 2023
GA20-19278S Corneal endothelial dystrophies - genetic causes and molecular mechanisms, Grant provider: The Czech Science Foundation; Main recipient: Charles University / 1st Medicine faculty; Recipient: prof. MUDr. Petra Lišková, MD, Ph.D., Period of research project: 2020 – 2023
NU20-07-00182 Childhood-onset retinal dystrophies; multidisciplinary management with novel approaches to accelerate molecular diagnosis and to explore disease mechanisms, Grant provider: Ministry Of Health Of The Czech Republic, Main recipient: Charles University / 1st Medicine faculty; Recipient: prof. MUDr. Petra Lišková, MD, Ph.D., Period of research project: 2020 – 2023
NV19-08-00122 Improved Interpretation of Next Generation Sequencing Datasets in Patients with Inherited Cardiomyopathy Using Functional Models Based on Induced Pluripotent Stem Cells and Advanced Cardiac Imaging in Families, Grant provider: Ministry Of Health Of The Czech Republic, Main recipient: Charles University / 1st Medicine faculty; Recipient: RNDr. Robert Dobrovolný, Ph.D., Period of research project: 2019 – 2023
NV19-08-00137 The application of new methods of genomic analysis in cases of rare genetic based diseases with negative results of genetic and genomic analyses. Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: Mgr. Viktor Stránecký, PhD; Period of research project: 2019 - 2023
NU20-04-00332 Organic and functional comorbidities in functional movement disorders, common pathophysiological mechanisms, biomarkers and prognostic factors, Grant provider: Ministry Of Health Of The Czech Republic, Main recipient: Charles University / 1st Medicine faculty; Recipient: MUDr. Tereza Serranová, Ph.D., Period of research project: 2020 – 2023
LM2018132 The National Center for Medical Genomic, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University in Prague / First Faculty of Medicine, Recipient: Prof. Ing. Stanislav Kmoch, CSc. Period of research project: 2020 - 2022
8F20004 Solving missing heritability in inherited retinal diseases using integrated omics and gene editing in human cellular and animal models, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University / 1st Medicine faculty; Recipient: doc. MUDr. Petra Lišková, Ph.D., Period of research project: 2020 – 2022
EF18_046/0015515 Modernization and instrumental upgrade of the National Center for Medical Genomics, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University in Prague / First Faculty of Medicine, Recipient: Prof. Ing. Stanislav Kmoch, CSc. Period of research project: 2020 - 2021
LTAUSA19068 Advancing the diagnosis, understanding, and treatment of inherited kidney disease related to mucin-1 through improved family identification and new genetic, molecular, and biochemical approaches, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University in Prague / First Faculty of Medicine, Recipient: prof. Ing. Stanislav Kmoch, CSc. Period of research project: 2019 - 2022
8F19002 Towards a new era for the identification and characterisation of inborn errors of glycosylation, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University / 1st Medicine faculty; Recipient: RNDr. Hana Hansíková, CSc., Period of research project: 2019 – 2022
NV19-07-00136 Characterization of the molecular basis of rare genetic diseases of pediatric onset using new methods of genome analysis. Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: Prof. Ing. Stanislav Kmoch, CSc.; Period of research project: 2019 - 2022
779257 Solving the unsolved Rare Diseases — Solve-RD, Grant provider: Horizon 2020, H2020-SC1-2017-Single-Stage-RTD, Period of research project: 2018 – 2022
NV18-06-00032 Molecular basis and mechanisms of familial intrahepatic cholestasis, Grant provider: Ministry Of Health Of The Czech Republic, Main recipient: Institute for Clinical and Experimental Medicine; Recipient: : Prof. MUDr. Mgr. Milan Jirsa, CSc., Period of research project: 2018 – 2022
NV18-03-00024 Classification of variants of unknown significance in cancer susceptibility genes by the multiplex PCR/NGS-based analysis of their impact on pre-mRNA splicing, ,Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: MUDr. Petra Kleiblová, Ph.D. Period of research project: 2018 – 2021
NV17-30965A Mitochondrial disorders with instability of mitochondrial DNA, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: Ing. Markéta Tesařová, Ph.D., Period of research project: 2017 – 2021
17-29786A Identification and characterization of genetic factors contributing to inherited tubulointerstitial kidney disease. Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: Mgr. Martina Živná, Ph.D.; Period of research project: 2017-2020
NV17-30500A Congenital cataracts - next-generation sequencing in diagnosis and management, Grant provider: Ministry Of Health Of The Czech Republic, Main recipient: Charles University / 1st Medicine faculty; Recipient: prof. MUDr. Petra Lišková, MD, Ph.D., Period of research project: 2017 – 2020
EF16_013/0001634 National Centrum of Medical Genomics - modernization of infrastructure and research of genetic variation in the population, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University in Prague / First Faculty of Medicine, Recipient: Prof. Ing. Stanislav Kmoch, CSc. Period of research project: 2017 - 2019
LQ1604 BIOCEV: from Fundamental to Applied Research, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Institute of Molecular Genetics of the Czech Academy of Sciences, Recipient: doc. RNDr. Radislav Sedláček, Ph.D., Period of research project: 2016 – 2020
NV16-29959A Bioinformatics and functional analyses of susceptibility variants supporting the NGS-based testing of hereditary cancers in the Czech Republic, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient : : Prof. MUDr. Zdeněk Kleibl, Ph.D Period of research project: 2016 – 2019
NV15-27682A Next generation sequencing for early diagnosis and individualized treatment of dilated cardiomyopathy and related forms of cardiomyopathy, Grant provider: Ministry Of Health Of The Czech Republic, Main recipient: Institute for Clinical and Experimental Medicine; Recipient: MUDr. Miloš Kubánek, Ph.D., Period of research project: 2015 – 2019
NV15-27695A Characterization of genetic predisposition to ovarian cancer using Next Gene Sequencing analysis ,Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: MUDr. Petra Kleiblová, Ph.D. Period of research project: 2015 – 2018
NV15-28830A Identification of genetic factors contributing to hereditary breast cancer development and prognosis; Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 1st Medicine faculty; Recipient: Prof. MUDr. Zdeněk Kleibl, Ph.D Period of research project: 2015 – 2018
15-28208A Characterization of the molecular basis of rare genetic diseases of pediatric onset using new methods of genome analysis, Grant provider: MZ0 - Ministry Of Health Of The Czech Republic. Main recipient: Charles University in Prague / First Faculty of Medicine, Period of research project: 2015-2018.
GB14-36804G Center of mitochondrial biology and pathology (MITOCENTRUM), Grant provider: GA0 - The Czech Science Foundation (GA CR), Main recipient: Institute of Physiology CAS, Period of research project: 2014-2018.
GA14-21903S Genetic architecture of impulsive violence, Grant provider: GA0 - The Czech Science Foundation (GA CR), Main recipient: Charles University in Prague / First Faculty of Medicine, Period of research project: 2014-2016.
NT14025 Role of rare variants in genetic predisposition to statin myopathy, Grant provider: MZ0 - Ministry Of Health Of The Czech Republic, Main recipient: Institute for Clinical and Experimental Medicine, Period of research project: 2013-2015.
NT13116 Identification of the genetic and molecular basis of rare genetic disorders using novel genomic methods, Grant provider: MZ0 - Ministry Of Health Of The Czech Republic, Main recipient: Charles University in Prague / First Faculty of Medicine, Period of research project: 2012-2015.
LH12015 Identification and characterization of genetic factors contributing to chronic kidney disease, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Charles University in Prague / First Faculty of Medicine, Period of research project: 2012-2015.
International projects
313011ATA2 PROMEDICOV-19 Research into progressive methods for diagnosing COVID-19 and the biomarkers enabling early detection of individuals at increased risk of severe course of the disease, Grant provider: Ministry of transport and construction of the Slovak Republic, Ministry of Education, Science, Research and Sports of the Slovak Republic and the Research Agency; Project is co-financed by the EU; Recipient: MEDIREX GROUP ACADEMY n.o., Period of research project: 2020 – 2023
PP-COVID-20-0056 Creation of a system of early and rapid detection, identification and diagnosis of new infectious diseases with pandemic potential Research and Development Support Agency – pilot study COVID-19, Grant provider: Research and Development Support Agency, Recipient: MEDIREX GROUP ACADEMY n.o., Start of research project: 2020
B1MG, 951724 Beyond 1M Genomes’ B1MG. Grant provider: European Union, Horizon2020; Period of research project: 2020-2023
831390 Orphanet Network — ONW. Grant provider: Public health EU DG Sante; Period of research project: 2018-2021
Second Faculty of Medicine at Charles University, Motol University Hospital
Solved projects
NU22-07-00165 - Identification and analysis of genes and genetic defects causing rare neurodevelopmental disorders in children; Grant provider Ministry of Health of the Czech Republic; Main recipient: Charles University Univerzita Karlova / 2nd Faculty of Medicine; Main recipient: prof. Ing. Zdeněk Sedláček DrSc; Period of research project: 2022 – 2025
ZD-ZDOVA2-001 Improvement of targetted prevention and early diagnostics in specific communicable and non-communicable diseases in selected socially excluded localities with Romani communities. Grant provider: Fonds Norway Grants a EHP; Main recipient: Motol University Hospital; Period of research project: 2021-2024
NU20-07-00049 Intestinal organoids as an in-vitro model for predicting the response to therapies correcting molecular defects in cystic fibrosis. Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 2nd Medicine faculty; Period of research project: 2020-2023
NV19-06-00189 Genetic factors of progressive hearing loss of cochlear implantation candidates: a potential for stratification of their therapy. Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Motol University Hospital; Period of research project: 2019-2022
NV19-06-00443 Genetics of adult-onset familial focal segmental glomerulosclerosis (FSGS) – implications for individualized therapy and successful kidney transplantation. Main recipient: Motol University Hospital; Period of research project: 2019-2022
NV18-02-00237 Detection of sudden cardiac death causes in people aged 0-35 years using molecular genetic methods – a pilot study. Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Charles University / 2nd Medicine faculty; Period of research project: 2018-2021
15-34904A Next generation sequencing in early Diagnosis and individualised therapy of hypertrophic cardiomyopathies, Grant provider: AZV ČR. Period of research project: 2015-2018.
LD14073 Introduction of next generation sequencing into clinical diagnostics in the Czech Republic: ethical and clinical genetic indication criteria, Grant provider: COST. Period of research project: 2014-2017.
CZ.2.16/3.1.00/24022 Improvement of early Diagnosis, prevention and therapy of severe reproductive-, prenatal and postnatal developmental disorders in children and adults: sustainability period, Grant provider: Operační program Praha Konkurenceschopnost. Period of research project: 2013-2018.
NF-CZ11-PDP-3-003-2014 National Coordinating Centre for rare diseases at the Motol University Hospital, Grant provider: Norway Grants. Period of research project: 2014-2017.
305444 RD-CONNECT, An integrated platform connecting registries, biobanks and clinical bioinformatics for rare disease research, Grant provider: FP7. Period of research project: 2012-2018.
Masaryk University – CEITEC
CZ.02.01.01/00/23_020/0008555 - Applicability of Comprehensive Genomic Testing – towards better diagnostics (ACGT2), Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT), Main recipient: Masaryk University in Brno; Project lead: prof. RNDr. Šárka Pospíšilová, Ph.D., Period of research project: 2025 – 2028
GA24-12028S - Constructing the Developmental Atlas of Alzheimer’s Disease using Cerebral Organoids and Single-Cell Transcriptomics; Grant provider: Czech Science Foundation; Main recipient: Masaryk university / Faculty of medicine; Other recipient: Institute of Biotechnology of the Czech Academy of Sciences; Project lead: Ing. Lukáš Valihrach Ph.D.; Period of research project: 2024 – 2026
Solved projects
NU22-03-00290 Study on exosomal microRNAs in gliomas: implications for diagnostics and innovative therapy, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC; Recipient: doc. Mgr. Jiří Šána, Ph.D., Period of research project: 2022 – 2025
GA22-35273S New approaches to Chimeric Antigen Receptor T cells: analysis of RASAL3 protein function and identification of novel CAR-T activity-modifying genes, Grant provider GA0 – Czech Science Foundation; Main recipient: Masaryk University / CEITEC; Recipient: doc. Michal Šmída, PhD., Period of research project: 2022 – 2024
NU21-04-00305 Transcriptomics and DNA methylation analysis in patients with focal cortical dysplasia, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / Medicine Faculty; Recipient: MUDr. Milan Brázdil, PhD., Period of research project: 2021 – 2024
NU20-03-00240 Whole exome, low-coverage genome and transcriptome sequencing as tools for precision oncology in paediatric patients with high-risk and relapsed solid tumors, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / Medicine Faculty; Recipient: prof. MUDr. Jaroslav Štěrba, Ph.D., Period of research project: 2020 – 2023
NU20-08-00137 Searching and functional testing of gene variants predisposing to familial haematopoietic disorders, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC; Recipient: Prof. MUDr. Michael Doubek, Ph.D., Period of research project: 2020 – 2023
NU20-08-00314 Single cell analysis: a modern tool to study clonal evolution in high-risk patients with chronic lymphocytic leukemia, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC; Recipient: prof. RNDr. Šárka Pospíšilová, Ph.D., Period of research project: 2020 – 2023
NV19-03-00562 Global methylation profiles and comprehensive analysis of gene fusions as a basis for therapeutic planning in pediatric oncology, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / Medicine Faculty; Recipient: prof. MUDr. Jaroslav Štěrba, Ph.D., Period of research project: 2019 – 2023
NV19-03-00501 Study of PIWI-interacting RNAs in glioblastoma stem cells and their potential clinical implications, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC; Recipient: doc. Mgr. Jiří Šána, Ph.D., Period of research project: 2019 – 2023
EF16_026/0008448 Analysis of Czech Genomes for Theranostics, Grant provider: The Ministry of Education, Youth and Sports (MEYS, MŠMT) ; Main recipient: Masaryk University / CEITEC; Recipient: prof. RNDr. Šárka Pospíšilová, Ph.D., Period of research project: 2019 – 2023
GA19-20873S Driver mutations in cancer genome of Nothobranchius furzeri: from tumor biology to concept of experimental model of spontaneous carcinogenesis, Grant provider GA0 – Czech Science Foundation; Main recipient: Masaryk University / CEITEC; Recipient: prof. RNDr. Ondřej Slabý, Ph.D., Period of research project: 2019 – 2023
NV19-03-00091 Comprehensive prognostic and predictive panel for chronic lymphocytic leukemia: a next-generation sequencing tool suitable for clinical practice and study of genetic architecture behind the disease evolution, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC; Recipient: prof. RNDr. Šárka Pospíšilová, Ph.D., Period of research project: 2019 – 2022
GA19-11931S The role of miRNA in development of glutamatergic and GABAergic signaling after early-life seizures, Grant provider GA0 – Czech Science Foundation; Main recipient: Masaryk University / CEITEC; Recipient: prof. MUDr. Milan Brázdil, Ph.D., Period of research project: 2019 – 2022
TJ02000049 Development and validation of innovative detection systems for quantification of urinary microRNAs in diagnosis of urologic cancers, Grant provider TA0 Technologic agentury of the Czech Republic; Main recipient: Masaryk University / CEITEC; Recipient: Mgr. Jiří Šána, Ph.D., Period of research project: 2019 – 2021
NV18-03-00054 THE ROLE OF MICRORNAS AND THEIR TARGETS IN THE TRANSFORMATION OF FOLLICULAR LYMPHOMA AND AGGRESSIVENESS OF CHRONIC LYMPHOCYTIC LEUKEMIA, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC; Recipient: Doc. MUDr. Mgr. Marek Mráz, Ph.D., Period of research project: 2018 – 2021
GA16-18257S Uncovering the mechanism underlying the tumor-suppressive effects of miR-215 and its substitution as new therapeutic strategy in colorectal cancer, Grant provider GA0 – Czech Science Foundation; Main recipient: Masaryk University / CEITEC; Recipient: prof. RNDr. Ondřej Slabý, Ph.D., Period of research project: 2016 – 2020
GA16-04726S miRNA and biochemical pathways in epilepsy, Grant provider GA0 – Czech Science Foundation; Main recipient: Masaryk University / CEITEC; Recipient: prof. MUDr. Milan Brázdil, Ph.D.,Period of research project: 2016 – 2020
NV16-34414A Determination of genetic regions susceptible to splicing affecting mutations, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC, Recipient: prof. MUDr. Tomáš Freiberger, Ph.D., Period of research project: 2016 – 2019
NV16-33209A Genome-wide expression profiling and mutation analysis as the diagnostic basis for personalized pediatric cancer treatment plans: a feasibility study, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / Medicine faculty, Recipient: prof. MUDr. Jaroslav Štěrba, Ph.D., Period of research project: 2016 – 2019
NV16-29447A Searching for mutations predisposing to familial hematologic and oncologic diseases), Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC, Recipient: Prof. MUDr. Michael Doubek, Ph.D., Period of research project: 2016 – 2019
NV15-31834A Selection of genomic defects in chronic lymphocytic leukemia and their impact on disease outcome, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC, Recipient: prof. RNDr. Šárka Pospíšilová, Ph.D., Period of research project: 2015 – 2018
NV15-33561A Resistance to monoclonal antibody therapy at B-CLL and B-lymphomas: its foundations and potential intervention strategies, Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: Masaryk University / CEITEC, Recipient: Mgr. Michal Šmída, Ph.D., Period of research project: 2015 – 2018
Project NGS-PTL - Next Generation Sequencing Platform for Targeted Personalized Therapy of Leukemia, FP7, number 306242 – the project goal is the application of the most modern approaches for genomics analysis for identification of novel diagnostics and prognostics biomarkers of leukemia and subsequent patient stratification and more effective application of therapeutical methodologies.
Projekt ALKATRAS (ALK activation as a target of translational science: break free from cancer, H2020, číslo 675712 ) – its main aim is to clarify yet unknown mechanisms of ALK-induced tumorigenesis and describe more specific role of oncogene ALK (anaplastic lymphoma kinase) in oncologic diseases like lymphomas, neuroblastomas and non-cell lung carcinoma (NSCLC).
University Hospital Brno
EF16_019/0000868 Molecular, cellular and clinical approach to healthy ageing. Grant provider: Ministry Of Health Of The Czech Republic, Main recipient St. Anne's University Hospital in Brno; Recipient: Dr. Gorazd B. Stokin, Period of research project: 2018 – 2023
NV15-30015A Analysis of clonal heterogeneity in chronic lymphocytic leukemia using next generation sequencing of B cell receptor. A national study. Grant provider: Ministry Of Health Of The Czech Republic; Main recipient: St. Anne's University Hospital in Brno, Recipient: Prof. MUDr. Michael Doubek, Ph.D., Period of research project: 2015 – 2018
Project supported by Agency of biomedical research (AZV, number 15-30015A ) entitled as „Analysis of clonal heterogeneity of chronic lymphocytic leukemia using next-generation sequencing of B-cell receptor gene. National study.“ has an ambitious aim to establish technology for high throughput sequencing of immunoglobulin genes and perform national screening of newly diagnosed CLL patients.
New-generation sequencing is used as a tool within the project entitled „Generation and development of genetic defects in leukemia“ (AZV, number 15-10035S) for identification of genetic disorders contributing to clonal evolution of leukemia and for study of B-cell receptor mediated signalling that both play important role in cellular response to external stimuli and thus contribute to clonal evolution.
Institute of Molecular and Translational Medicine - Olomouc
Information about current projects: https://imtm.cz/featured-research-projects.
VI20202022123 Epigenetic age of blood or sperm trace donor. Grant provider: Ministry of the Interior of the Czech Republic, Main recipient: Palacký University in Olomouc; Recipients: prof. Mgr. Jiří Drábek, Ph.D. a Mgr. Vilém Skyba; Period of research project: 2020-2022
MPO 67986/17/61600/3563 ENIGMA CZ - Czech National Genomic Map; Grant provider: Ministry of Industry and Trade of the Czech Republic, Main recipient: Institute of Applied Biotechnologies a.s., Period of research project: 2017-2022
ÚMTM addresses currently a number of international (EATRIS, FP7 INFLACARE, BBMRI etc.) and national grant projects (TACR, GACR, AZV etc.) focused on translational medicine, see more imtm.cz.
Available technologies
Institutes of First and Second Faculty of Medicine at Charles University in Prague
Sample Preparation and Laboratory Automation
- Covaris E220 Focused Ultrasonicator
- Tecan Freedom EVO® 150 Pipetting Robot
NGS Sequencing Technologies
- AVITI24 NextGen Multiomics Platform
- Illumina NovaSeq X
- Oxford Nanopore PromethION 2 Solo sequencer
- Oxford Nanopore MiniON long read sequencer
- ABI 3500xL Capillary Sequencers
PCR Technologies
- Bio-Rad CFX96 Touch Real-Time PCR System
- Applied Biosystems 7900HT Fast Real-Time PCR System
- Bio-Rad C1000 and T100 Thermal Cyclers
Supporting Infrastructure and Expertise
- Comprehensive equipment and expertise for sample preparation and bioinformatics data analysis
- Data storage and high-performance computing infrastructure
- Instrument specialists and bioinformaticians available for the infrastructure operation and data analysis


CEITEC MU Brno
- Massive paralel sequencers (Illumina MiSeq and Illumina NextSeq 500, MGI)
- Nanopore sequencing (Oxford Nanopore MinION a PromethION)
- Real-time qPCR and digital PCR (Life Technologies QuantStudio 12k, Life Technologies BioRad QX200, Roche LC480)
- System for single-cell analysis (10X Chromium)
- Automated pipetting and isolation Robot (TECAN FreedomEvo, Opentrons OT-2, RBC MagCore)
- Redudant data storage and computational servers (cooperation with eINFRA a ELIXIR-CZ)
- Flow cytometer and sorter (BD, cooperation with EATRIS-CZ)
Link for the list of equpiment: https://cfg.ceitec.cz/equipment/
Department of Medical Genetics, Faculty of Medicine in Pilsen, Charles University Prague and the University Hospital in Pilsen
- Opentrons OT-2 robotic liquid handler
- QIAGEN QIAxcel automated capillary electrophoresis instrument
- Massive parallel sequencers (Life Technologies IonTorrent, Illumina MiSeq)
- Capillary sequencers (Life Technologies)
- Real-time qPCR (Corbett Research, Qiagen)
- Invitrogen Qubit Flex fluorometer
- Fluorescent microscopes (Olympus)
- Data storage and computational servers
- DNA/RNA amplification systems
Biomedical centre of the Faculty of Medicine in Pilsen
- Long-read sequencer PacBio Vega
- DNA microarray systems (Agilent, Innopsys)
Institute of Molecular and Translational Medicine
- Massively parallel sequencers (Illumina MiSeq and HiSeq Illumina, Qiagen PyroMark Q96, Roche GS Junior, Sequenom)
- Systems for automated preparation of sequencing libraries (Illumina Neoprep)
- Robotic system for nucleic acid isolation (Roche MagNA Pure 96 MagCore)
- Microarray system (Affymetrix)
- Real-Time qPCR (Roche LC 480, LC 1536, Cobas 4800, Eppendorf, BioRad CFX96)
- Data storage and computer servers
- The flow cytometer and sorter (BD FACS Aria Fusion II)
- Fluorescence microscope (Olympus BX60)
- Laser microdissection
NCMG Scientific Advisory Board
Dr. Robert Ivánek

Robert Ivánek received his doctoral degree in Molecular Biology and Genetics from the Charles University, Prague, Czech Republic in 2009. During last two years of his PhD studies he was running the Genomics Core facility at the Institute of Molecular Genetics, Prague, Czech Republic. From 2009 till 2012 he trained as postdoctoral fellow at Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland. In 2012 he became Head of Bioinformatics Core Facility at Department of Biomedicine (DBM), University of Basel, Basel Switzerland. In his scientific career he contributed to identification of molecular basis for several human rare diseases by applying novel combination of genomic tools and bioinformatic approaches. He also investigate the effect of sequence variation on transcription factor binding, DNA methylation and chromatin structure. His team at DBM provides bioinformatic expertise and training for analysis of high-throughput genomic data in biomedical field.
Dr. Jacek Majewski

Dr. Jacek Majewski obtained his Bachelor’s degree in physics and Master’s degree in electrical engineering from Stanford University, Palo Alto, USA in 1991. Following a short adventure in the real world, he returned to academics and in 1999 received a PhD. in Biological Sciences from Wesleyan University, USA. His post-doctoral training with Dr. Jurg Ott at the Rockefeller University in New York focused on statistical genetics and methodologies for identifying variants underlying human genetic disease. At the onset of the human genome era, his combined background in quantitative sciences, biology and statistics opened new venues into the field of genome sequence analysis. He is currently an Associate Professor at the Department of Human Genetics, McGill University, Montreal, Canada, Medical Scientist at the McGill University Health Centre, and Scientist at the Shriners Hospital. His research centers on the analysis of next generation sequencing data, with specific applications to human genetic disease and cancer research. He holds grants from major Canadian and US funding agencies and has co-authored over 220 publications.
Dr. Vladimír Beneš

Vladimír Beneš, Head of GeneCore, studied in Prague, the Czech Republic. He has been at EMBL since 1994 when he came as a postdoc to Ansorge group in the Biochemical Instrumentation Unit. Vladimir worked on development of methodology supporting genome-wide high-throughput sequencing, mainly in the sample processing part. In 2001 he was appointed to build EMBL Genomics Core Facility, a technology orientated service laboratory founded to assist researchers with functional genomics projects. This facility is currently utilizing mainly massively parallel sequencing & microarray technologies as well as qPCR. Among Vladimir’s tasks belong also assessment of new technologies and functional genomics applications, in particular their suitability for implementation in the environment of core facilities. He is also strongly involved in teaching of methods applied in this field.
Dr. Jiří Zavadil

Jiří Zavadil received his doctoral degree in Biomedicine/Molecular Genetics in 1998 from the Charles University, Prague, Czech Republic. From 1998 to 2003, he trained as postdoctoral fellow at Mount Sinai School of Medicine and at Albert Einstein College of Medicine, in New York. Between 2003 and 2012 he was Assistant/Associate Professor of Pathology, Director of the NYU Genome Technology Center and Faculty Member of the NYU Center for Health Informatics and Bioinformatics at the New York University Langone Medical Center, New York. In 2012, he became Head of the Molecular Mechanisms and Biomarkers Group at the WHO International Agency for Research on Cancer (IARC), Lyon, France. In his scientific career he has studied human pathologies including cancer, using innovative workflows and methodologies in genomics and bioinformatics. His work has been documented by dozens (of 110+ total) lead-authored or co-authored publications. His team at IARC focuses on deciphering molecular mechanisms of carcinogenesis, with a particular emphasis on understanding the effects of environmental causes of cancer, and on the impact of carcinogens on critical genome alterations leading to cancer cell phenotype outcomes.
Publications
Publications with the support of NCMG for the year 2025
- Ashraf R, Polasek-Sedlackova H, Marini V, et al (2025) RECQ4-MUS81 interaction contributes to telomere maintenance with implications to Rothmund-Thomson syndrome. Nat Commun 16:1302. https://doi.org/10.1038/s41467-025-56518-1
- Bartos M, Urik M, Buresova L, et al (2025) Bacteriome of the Middle Ear in Children and Young Adults With Cholesteatoma and Retraction Pocket: A Pilot Study. OTO open 9:e70051. https://doi.org/10.1002/oto2.70051
- Bosáková V, Papatheodorou I, Kafka F, et al (2025) Serotonin attenuates tumor necrosis factor-induced intestinal inflammation by interacting with human mucosal tissue. Exp Mol Med 57:364–378. https://doi.org/10.1038/s12276-025-01397-1
- Botka T, Smetanová S, Vinco A, et al (2025) Virion content unpacked by long-read sequencing: stress-induced changes in transmitted staphylococcal mobilome due to phage-satellite interactions. Nucleic Acids Res. 2025;53(20):gkaf1165. doi:10.1093/nar/gkaf1165
- Boudná M, Blavet N, Samoilenko T, et al (2025) Analysis of extracellular vesicles of frequently used colorectal cancer cell lines. BMC Cancer 25:555. https://doi.org/10.1186/s12885-025-13936-0
- Brichova M, Dlouha L, Tenglerova M, et al (2025) Oral, not gut microbiota diversity, reflects the inflammation and neoplasia in patients with uveitis and vitreoretinal lymphoma. J Ophthalmic Inflamm Infect. doi:10.1186/s12348-025-00517-2
- Cassani M, Niro F, Fernandes S, et al (2025) Regulation of Cell-Nanoparticle Interactions through Mechanobiology. Nano Lett. https://doi.org/10.1021/acs.nanolett.4c04290
- Coufal S, Zakostelska ZJ, Thon T, et al (2025) Ocrelizumab transiently alters microbiota and modulates immune response depending on treatment outcome. iScience 28:. https://doi.org/10.1016/j.isci.2025.113872
- Černá M, Šťastná B, Pešek P, et al (2025) Technical Note: Semi-automated RNA Isolation from Tempus Blood RNA Tubes Using the Magcore Plus II Instrument. Folia Biol (Czech Republic) 71:88–94. https://doi.org/10.14712/fb2025071020088
- Deissova T, Tukmachi D Al, Radova L, et al (2025) Tissue MicroRNA Expression Signatures as Diagnostic Biomarkers and Predictors of Residual Disease Activity and Relapse in Treatment-Naïve Pediatric Inflammatory Bowel Disease. Inflamm Bowel.Dis 1–11
- Eid M, Bednaříková M, Vlažný J, et al (2025) Real‐World Data From a Molecular Tumor Board‐Assisted Cancer Care From a Single Center in The Czech Republic: Is Precision Oncology an Accessible Option, or a Privilege for a Minority of Patients? Cancer Med 14:. https://doi.org/10.1002/cam4.71119
- Engel C, Rendek M, Assoumani J, et al (2025) Comprehensive analysis of CNOT3-related neurodevelopmental disorders: phenotypic and genotypic characterization. Eur J Hum Genet 1–8. https://doi.org/10.1038/s41431-025-01884-z
- Fesiuk A, Pölöske D, de Araujo ED, et al (2025) Thyroid hormone receptor beta signaling is a targetable driver of prostate cancer growth. Mol Cancer 24:256. https://doi.org/10.1186/s12943-025-02451-2
- Giordano C, Kendler J, Sexl M, et al (2025) Anti-Cancer Potential of a new Derivative of Caffeic Acid Phenethyl Ester targeting the Centrosome. Redox Biol 81:. https://doi.org/10.1016/j.redox.2025.103582
- Helia O, Matúšová B, Havlová K, et al (2025) Chromosome engineering points to the cis-acting mechanism of chromosome arm-specific telomere length setting and robustness of plant phenotype, chromatin structure and gene expression. Plant J 121:e70024. https://doi.org/10.1111/tpj.70024
- Hrala M, Deissová T, Andrla P, et al (2025) Tissue and stool microbiome in pediatric inflammatory bowel disease patients: diversity differs in patients with relapsing and non-relapsing Crohn’s disease. Gut Pathog 17:90. https://doi.org/10.1186/s13099-025-00766-5
- Knotek P, Pusztai M, Jadrná I, Škaloud P (2025) Expanding our knowledge of Synurales (Chrysophyceae) in Florida, United States: Proposing three novel Mallomonas taxa. J Phycol 1–19. https://doi.org/10.1111/jpy.70062
- Kreisinger J, Kaňková Š, Dlouhá D, et al (2025) Associations between psychological or biological stress indicators and gut microbiota in pregnant women – findings from a prospective longitudinal study. BMC Microbiol 25:. https://doi.org/10.1186/s12866-025-04146-6
- Kubovčiak J, Kreisinger J (2025) Host Diet Preference Drives Diversity and Composition of Gut Microbiota in Captive Birds. Ecol Evol. doi:10.1002/ece3.72463
- Kumar J, Micka M, Komárek J, et al (2025) A class III ligand oscillates between internal and terminal binding modes as it engages with the Dishevelled PDZ domain. Structure. https://doi.org/https://doi.org/10.1016/j.str.2025.05.012
- Kusová A, Holá M, Petrová IG, et al (2025) TRB proteins in moss reveal their evolutionarily conserved roles in plant development and telomere maintenance. Plant J. 2025;124(3):e70574. doi:10.1111/tpj.70574
- Mašlaňová I, Kovařovic V, Botka T, et al (2025) Evidence of in vitro mecB-mediated β-lactam antibiotic resistance transfer to Staphylococcus aureus from Macrococcus psychrotolerans sp. nov., a psychrophilic bacterium from food-producing animals and human clinical specimens. Appl Environ Microbiol e0165224. https://doi.org/10.1128/aem.01652-24
- Mušálková D, Radina M, Kidd K, et al (2025) Plasma Metabolites Associated with CKD Stage in Autosomal Dominant Tubulointerstitial Kidney Disease. Kidney360 10–34067
- Navrkalova V, Mareckova A, Hricko S, et al (2025) Reliable detection of CNS lymphoma-derived circulating tumor DNA in cerebrospinal fluid using multi-biomarker NGS profiling: insights from a real-world study. Biomark Res 13:1–12. https://doi.org/10.1186/s40364-025-00777-z
- Navrkalova V, Mareckova A, Porc J, et al (2025) Advanced NGS analysis of cell-free tumor DNA supports clonal relation to primary high-grade B-cell lymphoma lesion and CNS relapse despite MRI negativity. Diagn Pathol 20:14. https://doi.org/10.1186/s13000-025-01609-2
- Papatheodorou I, Blažková G, Bosáková V, et al (2025) Redefining the role of IL-18 in post-surgical recovery and sepsis: a key mediator of inflammation resolution. J Transl Med 23:. https://doi.org/10.1186/s12967-025-06652-7
- Pavlova S, Malcikova J, Radova L, et al (2025) Detection of clinically relevant variants in the TP53 gene below 10% allelic frequency: A multicenter study by ERIC, the European Research Initiative on CLL. HemaSphere 9:e70065. https://doi.org/10.1002/hem3.70065
- Pavlová Š, Svozilová H, Krzyžánková M, et al (2025) Hypomethylating agents increase L1 retroelement expression without inducing novel insertions in myeloid malignancies. Mol Oncol 1–12. https://doi.org/10.1002/1878-0261.70111
- Pivrncova E, Bohm J, Barton V, et al (2025) Viable bacterial communities in freshly pumped human milk and their changes during cold storage conditions. Int Breastfeed J 20:. https://doi.org/10.1186/s13006-025-00738-0
- Pokorna P, Orlickova J, Machackova T, et al (2025) Genomic complexity and evolution of high-grade serous ovarian cancer treated with platinum-based chemotherapy: advancing precision oncology beyond BRCA1/BRCA2. J Ovarian Res. https://doi.org/10.1186/s13048-025-01911-z
- Polešovská L, Trmačová S, Celiker C, et al (2025) Unveiling the cellular and molecular mechanisms of diabetic retinopathy with human retinal organoids. Cell Death Dis 16:892. https://doi.org/10.1038/s41419-025-08244-1
- Ruckova M, Al Tukmachi D, Vecera M, et al (2025) Circulating MicroRNAs do not provide a diagnostic benefit over tissue biopsy in patients with brain metastases. Sci Rep. https://doi.org/10.1038/s41598-025-31344-z
- Ryba L, Lerchovà T, Bronsky J, Vlčkovà M (2025) The role of exome data reanalysis in clarifying STXBP3 associated inflammatory bowel disease and hearing loss. Gastroenterol Rep 13:2024–2025. https://doi.org/10.1093/gastro/goaf053
- Sammut S, Gresova K, Tzimotoudis D, et al (2025) MiRBench: Novel benchmark datasets for microRNA binding site prediction that mitigate against prevalent microRNA frequency class bias. Bioinformatics 41:i542–i551. https://doi.org/10.1093/bioinformatics/btaf233
- Schott C, Alajmi M, Bukhari M, et al (2025) Genetic Testing in Adults over 50 Years with Chronic Kidney Disease: Diagnostic Yield and Clinical Implications in a Specialized Kidney Genetics Clinic. Genes. 2025; 16(4):408. https://doi.org/10.3390/genes16040408
- Schwarz M, Fišer M, Šodková L, et al (2025) Phenotype Analysis in Two Families With Otopalatodigital Syndrome Spectrum Disorder Based on FLNA Gene Variants. Clin Genet 1–6. https://doi.org/10.1111/cge.70056
- Sterba M, Pokorna P, Kyr M, et al (2025) NTRK2 fusion-driven neuroblastoma treated with targeted therapy: A case report. Pediatr Hematol Oncol J 10:100788. https://doi.org/https://doi.org/10.1016/j.phoj.2025.100788
- Stribna T, Peldova P, Vosecka T, et al (2025) Novel four-exon deletion in ryanodine receptor gene (RYR2) associated with mixed electric and structural cardiac phenotype. Europace 27:1–3. https://doi.org/10.1093/europace/euaf189
- Strych L, Zavoral T, Komrskova P, et al (2025) Two de novo UBR1 variants in trans as a cause of Johanson-Blizzard syndrome. Biomed Pap 169:98–106. https://doi.org/10.5507/bp.2025.005
- Thon T, Kopelentova E, Srutkova D, et al (2025) Skin and gut microbiota composition and immune regulatory response differentiate IgE and non-IgE cow’s milk allergy patients with atopic dermatitis. iScience. DOI: 10.1016/j.isci.2025.113943
- Tinka P, Pokorná P, Kýr M, et al (2025) Individualized therapeutic approaches for relapsed and refractory pediatric ependymomas: a single institution experience. J Neurooncol 173:479–488. https://doi.org/10.1007/s11060-025-05004-1
- Tolmaci B, Stejskal P, Zuffa P, et al (2025) Minimal residual disease and prognostic significance of circulating tumour cells in early-stage colorectal cancer. https://doi.org/10.5507/bp.2025.023
- Vyklicka K, Kucera J, Barton V, et al (2025) Effect of carbohydrate substrates on growth and enterotoxin gene expression in Bacillus cereus (pacificus). Sci Rep. https://doi.org/10.1038/s41598-025-31689-5
- Wayhelova M, Peldova P, Krebsova A, et al (2025) Intragenic TTN Deletions in a Single Family with Dilated Cardiomyopathy. Appl Clin Genet 18:211–217. https://doi.org/10.2147/TACG.S550190
- Zídková J, Lauerová B, Mensová L, et al (2025) Genetic and Structural Variations in Czech Patients With Congenital Myopathies. Clin Genet 1–6. https://doi.org/10.1111/cge.14782
- Zikanova M, Skopova V, Stuurman KE, et al (2025) Phosphoribosylformylglycinamidine Synthase (PFAS) Deficiency: Clinical, Genetic and Metabolic Characterisation of a Novel Defect in Purine de Novo Synthesis. J Inherit Metab Dis 48:1–8. https://doi.org/10.1002/jimd.70041
- Zimolova V, Burocziova M, Berkova L, et al (2025) Germline Jak2-R1063H mutation interferes with normal hematopoietic development and increases risk of thrombosis and leukemic transformation. Leukemia. https://doi.org/10.1038/s41375-025-02737-w
- Živná M, Dostálová G, Barešová V, et al (2025) Misprocessing of α-Galactosidase A, Endoplasmic Reticulum Stress, and the Unfolded Protein Response. J Am Soc Nephrol: 36(4):p 628-644, April 2025. | DOI: 10.1681/ASN.0000000535
Preprints (BioRxiv, MedRxiv):
- Anderson Jade K (2025) Androgens mediate sexual dimorphism in Pilarowski-Bjornsson Syndrome. medRxiv 1–60
- Celiker C, Hruba E, Kyriakou K, et al (2025) Photostimulation Improves Maturation of Human Photoreceptors. 5–10
- Coci EG, Gertzen CGW, Millan PC, et al (2025) DDX3X syndrome: a multicenter genotype-phenotype study
- Hnízda A, Martinez-Delgado B, Sanchez-Ponce D, et al (2025) De novo heterozygous variants in EHMT2 genocopy Kleefstra syndrome via loss of G9a methyltransferase activity. bioRxiv 2025.09.25.678439. https://doi.org/10.1101/2025.09.25.678439
- Hribkova H, Pospisilova V, Cerna KA, et al (2025) Viral Infection Induces Alzheimer’s Disease-Related Pathways and Senescence in iPSC-Derived Neuronal Models. bioRxiv. doi:10.1101/2025.06.11.659008
- Kusová A, Holá M, Goffová I, et al (2025) Molecular and Phenotypic Characterization of Telomere Repeat Binding ( TRBs ) Proteins in Moss : Evolutionary and Functional Perspectives. bioRxiv 1–33
- Raska J, Satkova M, Plesingrova K, et al (2025) The Alzheimer’s-Associated SORL1 p.Y1816C Variant Impairs APP Sorting, Axonal Trafficking, and Neuronal Activity in iPSC-Derived Brain Models. bioRxiv 2025.07.28.667160. https://doi.org/10.1101/2025.07.28.667160
- Szaraz D, Bohm J, Machacek C, et al (2025) Bacteriome-Based Oral Dysbiosis Index in Patients with Oral Squamous Cell Carcinoma DOI: https://doi.org/10.21203/rs.3.rs-7724435/v1
- Vrbacká A, Přistoupilová A, Kidd KO, et al (2025) Long-Read Sequencing of the MUC1 VNTR: Genomic Variation, Mutational Landscape, and Its Impact on ADTKD Diagnosis and Progression. bioRxiv 2025.09.06.673538. https://doi.org/10.1101/2025.09.06.673538
Theses with the support of NCMG for the year 2025
- HOVHANNISYAN, Milena. The role of individual genotypes in the development of germinal forms of solid tumors in adulthood (Podíl individuálních genotypů na vzniku germinálních forem solidních nádorů dospělého věku). Dissertation thesis, Charles University, 1st Faculty of the Medicine.
- Sobotková, Marta. Complement deficiencies with focus on C1 inhibitor deficiency (Poruchy v systému komplementu se zaměřením na deficit C1 inhibitoru). Dissertation thesis, Charles University, 2nd Faculty of the Medicine
Publications with the support of NCMG for the year 2024
- Amaratunga SA, Tayeb TH, Dusatkova P, et al (2024) High yield of monogenic short stature in children from Kurdistan, Iraq: a genetic testing algorithm for consanguineous families. Genet Med 101332. https://doi.org/10.1016/j.gim.2024.101332 (preproof)
- Aust AC, Vidova V, Coufalikova K, et al (2024) Fecal tryptophan metabolite profiling in newborns in relation to microbiota and antibiotic treatment. Appl Microbiol Biotechnol 108:504. https://doi.org/10.1007/s00253-024-13339-4
- Baumgartner D, Mušová Z, Zídková J, et al (2024) Genetic Landscape of Amyotrophic Lateral Sclerosis in Czech Patients. J Neuromuscul Dis 11:1035–1048. https://doi.org/10.3233/JND-230236
- Boudna M, Machackova T, Vychytilova-Faltejskova P, et al (2024) Investigation of long non-coding RNAs in extracellular vesicles from low-volume blood serum specimens of colorectal cancer patients. Clin Exp Med 24:1–14. https://doi.org/10.1007/s10238-024-01323-1
- Dudakova L, Noskova L, Kmoch S, et al (2024) Disruption of OVOL2 Distal Regulatory Elements as a Possible Mechanism Implicated in Corneal Endothelial Dystrophy. Hum Mutat 2024:. https://doi.org/10.1155/2024/4450082
- Eid M, Trizuljak J, Taslerova R, et al (2024) Incidental germline findings during comprehensive genomic profiling of pancreatic and colorectal cancer: single-centre, molecular tumour board experience. Mutagenesis 1–10. https://doi.org/10.1093/mutage/geae014
- Elhassan EAE, Kmochová T, Benson KA, et al (2024) A Novel Monoallelic ALG5 Variant Causing Late-onset ADPKD and Tubulointerstitial Fibrosis. Kidney Int Reports. https://doi.org/https://doi.org/10.1016/j.ekir.2024.04.031
- Franková V, Ješina P (2024) Genomic newborn screening – ethical questions and practical challenges. Ces Pediatr 79:349–352. https://doi.org/10.55095/CSPediatrie2024/025
- Furstova E, Drevinek P, Novotna S, et al (2024) Precision medicine in cystic fibrosis: predictive role of forskolin-induced swelling assay. Eur Respir J 2400156. https://doi.org/10.1183/13993003.00156-2024
- Hoferkova E, Seda V, Kadakova S, et al (2024) Stromal cells engineered to express T cell factors induce robust CLL cell proliferation in vitro and in PDX co-transplantations allowing the identification of RAF inhibitors as anti-proliferative drugs. Leukemia 38:1699–1711. https://doi.org/10.1038/s41375-024-02284-w
- Horackova K, Zemankova P, Nehasil P, et al (2024) A comprehensive analysis of germline predisposition to early-onset ovarian cancer. Sci Rep 14:1–12. https://doi.org/10.1038/s41598-024-66324-2
- Jugas R, Pokorna P, Adamcova S, et al (2024) Dataset of DNA methylation profiles of 189 pediatric central nervous system, soft tissue, and bone tumors. Data Br 55:110590. https://doi.org/https://doi.org/10.1016/j.dib.2024.110590
- Kasperova BJ, Mraz M, Svoboda P, et al (2024) Sodium-glucose cotransporter 2 inhibitors induce anti-inflammatory and anti-ferroptotic shift in epicardial adipose tissue of subjects with severe heart failure. Cardiovasc Diabetol 23:223. https://doi.org/10.1186/s12933-024-02298-9
- Kidd KO, Williams AH, Taylor A, et al (2024) Eight-fold increased COVID-19 mortality in autosomal dominant tubulointerstitial kidney disease due to MUC1 mutations: an observational study. BMC Nephrol 25:1–10. https://doi.org/10.1186/s12882-024-03896-1
- Klvanova E, Videnska P, Barton V, et al (2024) Resistome in the indoor dust samples from workplaces and households : a pilot study. Front Cell Infect Microbiol 1–10. https://doi.org/10.3389/fcimb.2024.1484100
- Kmochová T, Kidd KO, Orr A, et al (2024) Autosomal dominant ApoA4 mutations present as tubulointerstitial kidney disease with medullary amyloidosis. Kidney Int 105:799–811. https://doi.org/https://doi.org/10.1016/j.kint.2023.11.021
- Kopčilová J, Ptáčková H, Kramářová T, et al (2024) Large TRAPPC11 gene deletions as a cause of muscular dystrophy and their estimated genesis. J Med Genet 61:908 LP – 913. https://doi.org/10.1136/jmg-2024-110016
- Kováčová M, Hlaváč V, Koževnikovová R, et al (2024) Artificial Intelligence-Driven Prediction Revealed CFTR Associated with Therapy Outcome of Breast Cancer: A Feasibility Study. Oncology 1–12. https://doi.org/10.1159/000540395
- Kramárek M, Souček P, Réblová K, et al (2024) Splicing analysis of STAT3 tandem donor suggests non-canonical binding registers for U1 and U6 snRNAs. Nucleic Acids Res 1–16. https://doi.org/10.1093/nar/gkae147
- Kreisinger J, Dooley J, Singh K, et al (2024) Investigating the effects of radiation, T cell depletion, and bone marrow transplantation on murine gut microbiota. Front Microbiol 15:1–11. https://doi.org/10.3389/fmicb.2024.1324403
- Kubanek M, Binova J, Piherova L, et al (2024) Genotype is associated with left ventricular reverse remodelling and early events in recent-onset dilated cardiomyopathy. ESC Hear Fail 4127–4138. https://doi.org/10.1002/ehf2.15009
- Kurucova T, Reblova K, Janovska P, et al (2024) Unveiling the dynamics and molecular landscape of a rare chronic lymphocytic leukemia subpopulation driving refractoriness: insights from single-cell RNA sequencing. Mol Oncol 18:2541–2553. https://doi.org/10.1002/1878-0261.13663
- Lipovy B, Hladik M, Vyklicka K, et al (2024) Rare multi-fungal sepsis: a case of triple-impact immunoparalysis. Folia Microbiol (Praha) 69:903–911. https://doi.org/10.1007/s12223-024-01165-0
- Majed OAK, Majed FO, Almoamen NJ, et al (2024) Distribution of pathogenic variants in the CFTR gene in a representative cohort of people with cystic fibrosis in the Kingdom of Bahrain. Mol Genet Genomics 299:. https://doi.org/10.1007/s00438-024-02119-4
- Mušálková D, Přistoupilová A, Jedličková I, et al (2024) Increased burden of rare protein-truncating variants in constrained, brain-specific and synaptic genes in extremely impulsively violent males with antisocial personality disorder. Genes, Brain Behav 23:. https://doi.org/10.1111/gbb.12882
- Navrkalova V, Plevova K, Radova L, et al (2024) Integrative NGS testing reveals clonal dynamics of adverse genomic defects contributing to a natural progression in treatment-naïve CLL patients. Br J Haematol 204:240–249. https://doi.org/10.1111/bjh.19191
- Nemcova Y, Knotek P, Jadrná I, et al (2024) Nanopatterns on silica scales of Mallomonas (Chrysophyceae, Stramenopiles): Unraveling UV resistance potential and diverse response to UVA and UVB radiation. J Phycol 1256–1272. https://doi.org/10.1111/jpy.13496
- Nosková E, Svobodová V, Hypská V, et al (2024) High-throughput sequencing of Strongyloides stercoralis - a fatal disseminated infection in a dog. Parasitology. https://doi.org/10.1017/S0031182024000568
- Ondrisova L, Seda V, Hlavac K, et al (2024) FoxO1/Rictor axis induces a nongenetic adaptation to ibrutinib via Akt activation in chronic lymphocytic leukemia. J Clin Invest 134:. https://doi.org/10.1172/JCI173770
- Pafčo B, Nosková E, Ilík V, et al (2024) First insight into strongylid nematode diversity and anthelmintic treatment effectiveness in beef cattle in the Czech Republic explored by HTS metagenomics. Vet Parasitol Reg Stud Reports 47:. https://doi.org/10.1016/j.vprsr.2023.100961
- Palova H, Das A, Pokorna P, et al (2024) Precision immuno-oncology approach for four malignant tumors in siblings with constitutional mismatch repair deficiency syndrome. npj Precis Oncol 8:. https://doi.org/10.1038/s41698-024-00597-8
- Pecina P, Čunátová K, Kaplanová V, et al (2024) Haplotype variability in mitochondrial rRNA predisposes to metabolic syndrome. Commun Biol 7:. https://doi.org/10.1038/s42003-024-06819-w
- Peterková K, Konečný L, Macháček T, et al (2024) Winners vs. losers: Schistosoma mansoni intestinal and liver eggs exhibit striking differences in gene expression and immunogenicity. PLoS Pathog 20:1–25. https://doi.org/10.1371/journal.ppat.1012268
- Pivrncova E, Buresova L, Kotaskova I, et al (2024) Impact of intrapartum antibiotic prophylaxis on the oral and fecal bacteriomes of children in the first week of life. Sci Rep 14:1–17. https://doi.org/10.1038/s41598-024-68953-z
- Poláčková P, Borovec J, Vašáková J, et al (2024) Using three-dimensional geometric morphometry for facial analysis in patients with the oculo-auriculo-vertebral spectrum. Orthod Craniofacial Res 917–927. https://doi.org/10.1111/ocr.12834
- Pospíšilová P, Čejková D, Buršiková P, et al (2024) The hare syphilis agent is related to, but distinct from, the treponeme causing rabbit syphilis. PLoS One 19:1–15. https://doi.org/10.1371/journal.pone.0307196
- Redmer T, Raigel M, Sternberg C, et al (2024) JUN mediates the senescence associated secretory phenotype and immune cell recruitment to prevent prostate cancer progression. Mol Cancer 23:1–21. https://doi.org/10.1186/s12943-024-02022-x
- Schmiedová L, Černá K, Li T, et al (2024) Bacterial communities along parrot digestive and respiratory tracts: the effects of sample type, species and time. Int Microbiol 27:127–142. https://doi.org/10.1007/s10123-023-00372-y
- Schwarz M, Gazdarica M, Froňková E, et al (2024) Functional studies associate novel DUOX2 gene variants detected in heterozygosity to Crohn’s disease. Mol Biol Rep 51:399. https://doi.org/10.1007/s11033-024-09317-8
- Schwarz M, Geryk J, Havlovicová M, et al (2024) Body mass index is an overlooked confounding factor in existing clustering studies of 3D facial scans of children with autism spectrum disorder. Sci Rep 14:1–12. https://doi.org/10.1038/s41598-024-60376-0
- Slaba K, Pokorna P, Jugas R, et al (2024) Diagnostic efficacy and clinical utility of whole-exome sequencing in Czech pediatric patients with rare and undiagnosed diseases. Sci Rep 14:1–11. https://doi.org/10.1038/s41598-024-79872-4
- Smolka V, Friedecky D, Kolarova J, et al (2024) Aminoacylase 1 deficiency: case report on three affected siblings. AME Case Reports 8:18–18. https://doi.org/10.21037/acr-23-46
- Spurná Z, Čapková P, Punová L, et al (2024) Clinical-genetic analysis of selected genes involved in the development of the human skeleton in 128 Czech patients with suspected congenital skeletal abnormalities. Gene 892:. https://doi.org/10.1016/j.gene.2023.147881
- Stork M, Ondrouskova E, Bohunova M, et al (2024) Del(1p32) is an early and high-risk event in multiple myeloma patients with extraosseous disease. Blood Cancer J 14:6–8. https://doi.org/10.1038/s41408-024-01131-6
- Strych L, Černá M, Hejnalová M, et al (2024) Targeted long-read sequencing identified a causal structural variant in X-linked nephrogenic diabetes insipidus. BMC Med Genomics 17:1–7. https://doi.org/10.1186/s12920-024-01801-1
- Svozilova H, Vojtova L, Matulova J, et al (2024) In vitro culture of leukemic cells in collagen scaffolds and carboxymethyl cellulose-polyethylene glycol gel. PeerJ 12:e18637. https://doi.org/10.7717/peerj.18637
- Šilhavý J, Mlejnek P, Šimáková M, et al (2023) Spontaneous nonsense mutation in the tuftelin 1 gene is associated with abnormal hair appearance and amelioration of glucose and lipid metabolism in the rat. Physiol Genomics 56:65–73. https://doi.org/10.1152/physiolgenomics.00084.2023
- Štika J, Pešová M, Kozubík KS, et al (2024) A novel thrombocytopenia-4-causing CYCS gene variant decreases caspase activity: Three-generation study. Br J Haematol 1–9. https://doi.org/10.1111/bjh.19694
- Šubrt I, Zavoral T, Strych L, et al (2024) A recurrent synonymous L1CAM variant in a fetus with hydrocephalus. Hum Genome Var 11:1–4. https://doi.org/10.1038/s41439-024-00263-2
- Těšický M, Schmiedová L, Krajzingrová T, et al (2024) Nearly (?) sterile avian egg in a passerine bird. FEMS Microbiol Ecol 100:1–13. https://doi.org/10.1093/femsec/fiad164
- Trizuljak J, Likavcová P, Staňo Kozubík K, et al (2024) Impact of thrombocytopenia-associated c.-118C>T and c.-140C>G ANKRD26 5’UTR variants in three-generational pedigree. Platelets 35:2388103. https://doi.org/10.1080/09537104.2024.2388103
- Vymazal O, Papatheodorou I, Andrejčinová I, et al (2024) Calcineurin-NFAT signaling controls neutrophils’ ability of chemoattraction upon fungal infection. J Leukoc Biol 3:816–829. https://doi.org/10.1093/jleuko/qiae091
- Zakostelska Jiraskova Z, Kraus M, Coufal S, et al (2024) Lysate of Parabacteroides distasonis prevents severe forms of experimental autoimmune encephalomyelitis by modulating the priming of T cell response. Front Immunol 15:1475126. https://doi.org/10.3389/fimmu.2024.1475126
- Zemankova P, Cerna M, Horackova K, et al (2024) A deep intronic recurrent CHEK2 variant c.1009-118_1009-87delinsC affects pre-mRNA splicing and contributes to hereditary breast cancer predisposition. Breast 75:103721. https://doi.org/10.1016/j.breast.2024.103721
Theses with the support of NCMG for the year 2024
- Ebrahimi Naghani S (2024) Integrative phenotyping approaches to unmask the Phyb-PIF4 pathway in Arabidopsis thaliana reproductive organs at high ambient temperatures. – dissertation thesis, MUNI, FSc
- Jelínková S (2024) Charakterizace vrozených alterací genů podmiňujících vznik a prognózu dědičných forem vybraných nádorů dospělého věku (Characterization of germline alterations affecting genes influencing development and prognosis of specific adult cancers) – dissertation thesis, 1. LF UK, Biochemistry and patobiochemistry
- Remešová E (2024) Alternativy bisulfitační reakce pro detekci metylace cytosinu (Bisulfite reaction alternatives for cytosine methylation detection) - diploma theses, Univerzita Palackého v Olomouci, FSc
Publications with the support of NCMG for the year 2023
- Baird SJE, Hiadlovská Z, Daniszová K, et al (2023) A gene copy number arms race in action: X,Y-chromosome transmission distortion across a species barrier. Evolution (N Y) 77:1330–1340. https://doi.org/10.1093/evolut/qpad051
- Ballonová L, Souček P, Slanina P, et al (2023) Myeloid lineage cells evince distinct steady-state level of certain gene groups in dependence on hereditary angioedema severity. Front Genet 14:1–15. https://doi.org/10.3389/fgene.2023.1123914
- Bleyer AJ, Kidd KO, Williams AH, et al (2023) Maternal health and pregnancy outcomes in autosomal dominant tubulointerstitial kidney disease. Obstet Med 16:162–169. https://doi.org/10.1177/1753495X221133150
- Bohosova J, Kozelkova K, Al Tukmachi D, et al (2023) Long non-coding RNAs enable precise diagnosis and prediction of early relapse after nephrectomy in patients with renal cell carcinoma. J Cancer Res Clin Oncol 149:7587–7600. https://doi.org/10.1007/s00432-023-04700-7
- Bruel AL, Ganga AK, Nosková L, et al (2023) Pathogenic RAB34 variants impair primary cilium assembly and cause a novel oral-facial-digital syndrome. Hum Mol Genet 32:2822–2831. https://doi.org/10.1093/hmg/ddad109
- Cassani M, Fernandes S, Oliver-De La Cruz J, et al (2023) YAP Signaling Regulates the Cellular Uptake and Therapeutic Effect of Nanoparticles. Adv Sci 2302965:1–16. https://doi.org/10.1002/advs.202302965
- Celiker C, Weissova K, Cerna KA, et al (2023) Light-Responsive MicroRNA Molecules in Human Retinal Organoids are Differentially Regulated by Distinct Wavelengths of Light. iScience 26:107237. https://doi.org/10.1016/j.isci.2023.107237
- de Jesus AA, Chen G, Yang D, et al (2023) Constitutively active Lyn kinase causes a cutaneous small vessel vasculitis and liver fibrosis syndrome. Nat Commun 14:. https://doi.org/10.1038/s41467-023-36941-y
- Deissová T, Zapletalová M, Kunovský L, et al (2023) 16S rRNA gene primer choice impacts off-target amplification in human gastrointestinal tract biopsies and microbiome profiling. Sci Rep 13:1–10. https://doi.org/10.1038/s41598-023-39575-8
- Denommé-Pichon AS, Bruel AL, Duffourd Y, et al (2023) A Solve-RD ClinVar-based reanalysis of 1522 index cases from ERN-ITHACA reveals common pitfalls and misinterpretations in exome sequencing. Genet Med 25:. https://doi.org/10.1016/j.gim.2023.100018
- Fajkus P, Adámik M, Nelson ADL, et al (2023) Telomerase RNA in Hymenoptera (Insecta) switched to plant/ciliate-like biogenesis. Nucleic Acids Res 51:420–433. https://doi.org/10.1093/nar/gkac1202
- Fedorova V, Amruz Cerna K, Oppelt J, et al (2023) MicroRNA Profiling of Self-Renewing Human Neural Stem Cells Reveals Novel Sets of Differentially Expressed microRNAs During Neural Differentiation In Vitro. Stem Cell Rev Reports 1524–1539. https://doi.org/10.1007/s12015-023-10524-2
- Fedorova V, Pospisilova V, Vanova T, et al (2023) Glioblastoma and cerebral organoids : development and analysis of an in vitro model for glioblastoma migration. 17:647–663. https://doi.org/10.1002/1878-0261.13389
- Fulneček J, Klimentová E, Cairo A, et al (2023) The SAP domain of Ku facilitates its efficient loading onto DNA ends. Nucleic Acids Res 11706–11716. https://doi.org/10.1093/nar/gkad850
- Hejret V, Varadarajan NM, Klimentova E, et al (2023) Analysis of chimeric reads characterises the diverse targetome of AGO2-mediated regulation. Sci Rep 13:https://doi.org/10.1038/s41598-023-49757-z
- Hiatt SM, Trajkova S, Sebastiano MR, et al (2023) Deleterious, protein-altering variants in the transcriptional coregulator ZMYM3 in 27 individuals with a neurodevelopmental delay phenotype. Am J Hum Genet 110:215–227. https://doi.org/10.1016/j.ajhg.2022.12.007
- Ilík V, Kreisinger J, Modrý D, et al (2023) High diversity and sharing of strongylid nematodes in humans and great apes cohabiting an unprotected area in Cameroon. PLoS Negl Trop Dis 17:1–19. https://doi.org/10.1371/journal.pntd.0011499
- Janacova L, Stenckova M, Lapcik P, et al (2023) Catechol-O-methyl transferase suppresses cell invasion and interplays with MET signaling in estrogen dependent breast cancer. Sci Rep 13:1–14. https://doi.org/10.1038/s41598-023-28078-1
- Jorge S, Kidd K, Vylet’al P, et al (2023) Bi-allelic REN Mutations and Undetectable Plasma Renin Activity in a Patient With Progressive CKD. Kidney Int Reports 8:1112–1116. https://doi.org/10.1016/j.ekir.2023.01.017
- Kmochová T, Kidd KO, Orr A, et al (2023) Autosomal dominant ApoA4 mutations present as tubulointerstitial kidney disease with medullary amyloidosis. Kidney Int. https://doi.org/https://doi.org/10.1016/j.kint.2023.11.021
- Konecna E, Videnska P, Buresova L, et al (2023) Enrichment of human nasopharyngeal bacteriome with bacteria from dust after short-term exposure to indoor environment: a pilot study. BMC Microbiol 23:1–16. https://doi.org/10.1186/s12866-023-02951-5
- Kral J, Jelinkova S, Zemankova P, et al (2023) Germline multigene panel testing of patients with endometrial cancer. Oncol Lett 25:1–11. https://doi.org/10.3892/ol.2023.13802
- Kulkarni A, Jozefiaková J, Bhide K, et al (2023) Differential transcriptome response of blood brain barrier spheroids to neuroinvasive Neisseria and Borrelia. Front Cell Infect Microbiol 13:1–16. https://doi.org/10.3389/fcimb.2023.1326578
- Lapcik P, Sulc P, Janacova L, et al (2023) Desmocollin-1 is associated with pro-metastatic phenotype of luminal A breast cancer cells and is modulated by parthenolide. Cell Mol Biol Lett 28:. https://doi.org/10.1186/s11658-023-00481-6
- Mahrik L, Stefanovie B, Maresova A, et al (2023) The SAGA histone acetyltransferase module targets SMC5/6 to specific genes. Epigenetics and Chromatin 16:1–16. https://doi.org/10.1186/s13072-023-00480-z
- Mancikova V, Pesova M, Pavlova S, et al (2023) Distinct p53 phosphorylation patterns in chronic lymphocytic leukemia patients are reflected in the activation of circumjacent pathways upon DNA damage. Mol Oncol 17:82–97. https://doi.org/10.1002/1878-0261.13337
- Navrkalova V, Plevova K, Radova L, et al (2023) Integrative NGS testing reveals clonal dynamics of adverse genomic defects contributing to a natural progression in treatment-naïve CLL patients. Br J Haematol 1–10. https://doi.org/10.1111/bjh.19191
- Necpál J, Winkelmann J, Zech M, Jech R (2023) A de novo GRIA3 variant with complex hyperkinetic movement disorder in a girl with developmental delay and self-limited epilepsy. Park Relat Disord 111:. https://doi.org/10.1016/j.parkreldis.2023.105437
- Neřoldová M, Ciara E, Slatinská J, et al (2023) Exome sequencing reveals IFT172 variants in patients with non-syndromic cholestatic liver disease. PLoS One 18:e0288907. https://doi.org/10.1371/journal.pone.0288907
- Nosková L, Fukata Y, Stránecký V, et al (2023) ADAM22 ethnic-specific variant reducing binding of membrane-associated guanylate kinases causes focal epilepsy and behavioural disorder . Brain Commun 5:1–8. https://doi.org/10.1093/braincomms/fcad295
- Nosková, E., Modrý, D., Svobodová, V., Baláž, V., Červená, B., Dumas, L., ... & Pafčo, B. (2023). GENETIC DIVERSITY OF STRONGYLOIDES INFECTING DOGS. THE IMPACT OF GLOBAL CHANGE, 40.
- Pivovarcikova K, Pitra T, Alaghehbandan R, et al (2023) Lynch syndrome-associated upper tract urothelial carcinoma frequently occurs in patients older than 60 years: an opportunity to revisit urology clinical guidelines. Virchows Arch 483:517–526. https://doi.org/10.1007/s00428-023-03626-2
- Pivrncova E, Kotaskova I, Buresova L, et al (2023) THE EFFECT OF INTRAPARTUM ANTIBIOTIC PROPHYLAXIS ON NEONATAL GUT AND ORAL BACTERIOME. 65269705 (ppt)
- Pócsi M, Fejes Z, Bene Z, et al (2023) Human epididymis protein 4 (HE4) plasma concentration inversely correlates with the improvement of cystic fibrosis lung disease in p.Phe508del-CFTR homozygous cases treated with the CFTR modulator lumacaftor/ivacaftor combination. J Cyst Fibros 22:1085–1092. https://doi.org/10.1016/j.jcf.2023.04.001
- Redmer T, Raigel M, Sternberg C, Ziegler R (2023) JUN mediates senescence and immune cell recruitment to prevent prostate cancer progression (BioRxiv)
- Rehulkova A, Chudacek J, Prokopova A, et al (2023) Clinical and prognostic significance of detecting CEA, EGFR, LunX, c-met and EpCAM mRNA-positive cells in the peripheral blood, tumor-draining blood and bone marrow of non-small cell lung cancer patients. Transl Lung Cancer Res 12:1034–1050. https://doi.org/10.21037/tlcr-22-801
- Reiss Z, Rob F, Kolar M, et al (2023) Skin microbiota signature distinguishes IBD patients and reflects skin adverse events during anti-TNF therapy. Front Cell Infect Microbiol 12:1–15. https://doi.org/10.3389/fcimb.2022.1064537
- Roskova I, Vecera M, Radova L, et al (2023) Small RNA Sequencing Identifies a Six-MicroRNA Signature Enabling Classification of Brain Metastases According to their Origin. Cancer Genomics and Proteomics 20:18–29. https://doi.org/10.21873/cgp.20361
- Ruckova M, Tukmachi D Al Circulating MicroRNAs Do Not Provide a Diagnostic Benefit Over Tissue Biopsy in Patients With Brain Metastases. 1–23 (preprint)
- Schmiedová L, Černá K, Li T, et al (2023) Bacterial communities along parrot digestive and respiratory tracts: the effects of sample type, species and time. Int Microbiol. https://doi.org/10.1007/s10123-023-00372-y
- Schwarz M, Kanich O, Berka M, Havlovicová M, Drahanský M. Dermatoglyphic Patterns in Monozygotic Twins with Zimmermann-Laband Syndrome. Journal of Forensic Identification. 2023;73(1)
- Silhavy J, Mlejnek P, Simakova M, et al (2023) Spontaneous nonsense mutation in Tuft1 (tuftelin 1) gene is associated with abnormal hair appearance and amelioration of glucose and lipid metabolism in the rat. Physiol Genomics 65–73. https://doi.org/10.1152/physiolgenomics.00084.2023
- Skopkova M, Stufkova H, Rambani V, et al (2023) ATAD3A-related pontocerebellar hypoplasia: new patients and insights into phenotypic variability. Orphanet J Rare Dis 18:1–12. https://doi.org/10.1186/s13023-023-02689-3
- Sterba M, Pokorna P, Faberova R, et al (2023) Targeted treatment of severe vascular malformations harboring PIK3CA and TEK mutations with alpelisib is highly effective with limited toxicity. Sci Rep 13:1–9. https://doi.org/10.1038/s41598-023-37468-4
- Těšický M, Schmiedová L, Krajzingrová T, et al. Nearly (?) sterile avian egg in a passerine bird. FEMS Microbiology Ecology. 2023:fiad164.
- Vaculík O, Chalupová E, Grešová K, et al (2023) Transfer Learning Allows Accurate RBP Target Site Prediction with Limited Sample Sizes. Biology (Basel) 12:1276. https://doi.org/10.3390/biology12101276
- Vanova T, Sedmik J, Raska J, et al (2023) Cerebral organoids derived from patients with Alzheimer’s disease with PSEN1/2 mutations have defective tissue patterning and altered development. Cell Rep 42:113310. https://doi.org/10.1016/j.celrep.2023.113310
- Vecera L, Prasil P, Srovnal J, et al (2023) Morphine Analgesia, Cannabinoid Receptor 2, and Opioid Growth Factor Receptor Cancer Tissue Expression Improve Survival after Pancreatic Cancer Surgery. Cancers (Basel) 15:. https://doi.org/10.3390/cancers15164038
- Vejmelkova K, Pokorna P, Noskova K, et al (2023) Tazemetostat in the therapy of pediatric INI1-negative malignant rhabdoid tumors. Sci Rep 13:1–5. https://doi.org/10.1038/s41598-023-48774-2
- Vidlarova M, Rehulkova A, Stejskal P, et al (2023) Recent Advances in Methods for Circulating Tumor Cell Detection. Int J Mol Sci 24:. https://doi.org/10.3390/ijms24043902
- Waterham HR, Koster J, Ebberink MS, et al (2023) Autosomal dominant Zellweger spectrum disorder caused by de novo variants in PEX14 gene. Genet Med 25:. https://doi.org/10.1016/j.gim.2023.100944
- Zídková J, Kramářová T, Kopčilová J, et al (2023) Genetic findings in Czech patients with limb girdle muscular dystrophy. Clin Genet 104:542–553. https://doi.org/10.1111/cge.14407
- Zunova H, Stolfa M, Kunikova T, et al (2023) A unique coincidence of a 17q12 deletion and duplication in a Czech family led to a refined genotype–phenotype correlation. Am J Med Genet Part A. https://doi.org/10.1002/ajmg.a.63085
Theses with the support of NCMG for the year 2023
- Král (2023) Identifikace dědičných faktorů ovlivňujících vznik karcinomu pankreatu a dalších solidních tumorů, disertation theses, Charles University, 1st Faculty of Medicine
- Rada M (2023) Analýza vybraných orálních probiotik z kultur a z orálního prostředí – bachelor thesis, MU Brno
- Těšický M (2023) Host-microbiota , pro-inflammatory immunity and physiological senescence in wild birds – disertation theses, Charles University, Faculty of Science
- Zůnová M (2023) Analýza genomických variant u pacientů s neurovývojovými onemocněními se zaměřením na epilepsie – disertation theses, Charles University, 2nd Faculty of Medicine
Publications with the support of NCMG for the year 2022
- Arpas T, Jelinkova H, Hrabovsky S, et al (2022) Very rare near‐haploid acute lymphoblastic leukemia resistant to immunotherapy and CAR‐T therapy in 19‐year‐old male patient. Clin Case Reports 10:. https://doi.org/10.1002/ccr3.5545
- Baloun J, Pekacova A, Wenchich L, et al (2022) Menopausal Transition: Prospective Study of Estrogen Status, Circulating MicroRNAs, and Biomarkers of Bone Metabolism. Front Endocrinol (Lausanne) 13:. https://doi.org/10.3389/fendo.2022.864299
- Bhide K, Mochnáčová E, Tkáčová Z, et al (2022) Signaling events evoked by domain III of envelop glycoprotein of tick-borne encephalitis virus and West Nile virus in human brain microvascular endothelial cells. Sci Rep 12:. https://doi.org/10.1038/s41598-022-13043-1
- Bleyer AJ, Wolf MT, Kidd KO, et al (2022) Autosomal dominant tubulointerstitial kidney disease: more than just HNF1β. Pediatr Nephrol. doi: 10.1007/s00467-021-05118-4.
- Bleyer AJ, Kidd KO, Williams AH, et al (2022) Maternal health and pregnancy outcomes in autosomal dominant tubulointerstitial kidney disease. Obstetric Medicine. 0(0). doi:10.1177/1753495X221133150
- Bohosova J, Kasik M, Kubickova A, et al (2022) LncRNA PVT1 is increased in renal cell carcinoma and affects viability and migration in vitro. J Clin Lab Anal 36:. https://doi.org/10.1002/jcla.24442
- Boublikova L, Kramarzova KS, Zwyrtkova M, et al (2022) The clinical value of circulating free tumor DNA in testicular germ cell tumor patients. Urol Oncol Semin Orig Investig 40:412.e15-412.e24. https://doi.org/10.1016/j.urolonc.2022.04.021
- Cediel ML, Stawarski M, Blanc X, et al (2022) GABBR1 monoallelic de novo variants linked to neurodevelopmental delay and epilepsy. Am J Hum Genet 109:1885–1893. https://doi.org/10.1016/j.ajhg.2022.08.010
- Costa A, Franková V, Robert G, et al (2022) Co-designing models for the communication of genomic results for rare diseases: a comparative study in the Czech Republic and the United Kingdom. J Community Genet 13:313–327. https://doi.org/10.1007/s12687-022-00589-w
- Dolina J, Kunovsky L, Kroupa R, et al (2022) Achalasia and acromegaly: Co-incidence of these diseases or a new syndrome? Biomed Pap 166:228–235. https://doi.org/10.5507/bp.2021.040
- Elhassan EAE, Murray SL, Connaughton DM, et al (2022) The utility of a genetic kidney disease clinic employing a broad range of genomic testing platforms: experience of the Irish Kidney Gene Project. J Nephrol 35:1655–1665. https://doi.org/10.1007/s40620-021-01236-2
- Ergir E, Oliver-De La Cruz J, Fernandes S, et al (2022) Generation and maturation of human iPSC-derived 3D organotypic cardiac microtissues in long-term culture. Sci Rep 12:. https://doi.org/10.1038/s41598-022-22225-w
- Fajkus P, Adámik M, Nelson ADL, et al (2022) Telomerase RNA in Hymenoptera (Insecta) switched to plant/ciliate-like biogenesis. Nucleic Acids Res. https://doi.org/10.1093/nar/gkac1202
- Finstrlová A, Mašlaňová I, Blasdel Reuter BG, et al (2022) Global Transcriptomic Analysis of Bacteriophage-Host Interactions between a Kayvirus Therapeutic Phage and Staphylococcus aureus. Microbiol Spectr 10:. https://doi.org/10.1128/spectrum.00123-22
- Guizzo MG, Dolezelikova K, Neupane S, et al (2022) Characterization and manipulation of the bacterial community in the midgut of Ixodes ricinus. Parasites and Vectors 15:. https://doi.org/10.1186/s13071-022-05362-z
- Hrbac T, Kopkova A, Siegl F, et al (2022) HLA-E and HLA-F Are Overexpressed in Glioblastoma and HLA-E Increased After Exposure to Ionizing Radiation. Cancer Genomics and Proteomics 19:151–162. https://doi.org/10.21873/cgp.20311
- Jaroušek R, Mikulová A, Daďová P, et al (2022) Single-cell RNA sequencing analysis of T helper cell differentiation and heterogeneity. Biochim Biophys Acta - Mol Cell Res 1869:. https://doi.org/10.1016/j.bbamcr.2022.119321
- Limberger T, Schlederer M, Trachtová K, et al (2022) KMT2C methyltransferase domain regulated INK4A expression suppresses prostate cancer metastasis. Mol Cancer 21:. https://doi.org/10.1186/s12943-022-01542-8
- Macečková Z, Kubíčková A, De Sanctis JB, Hajdúch M (2022) Effect of Glucocorticosteroids in Diamond-Blackfan Anaemia: Maybe Not as Elusive as It Seems. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23031886
- Mancikova V, Pesova M, Pavlova S, et al (2022) Distinct p53 phosphorylation patterns in chronic lymphocytic leukemia patients are reflected in the activation of circumjacent pathways upon DNA damage. Mol Oncol. https://doi.org/10.1002/1878-0261.13337
- Martino F, Varadarajan NM, Perestrelo AR, et al (2022) The mechanical regulation of RNA binding protein hnRNPC in the failing heart
- Mason B, Petrzelkova KJ, Kreisinger J, et al (2022) Gastrointestinal symbiont diversity in wild gorilla: A comparison of bacterial and strongylid communities across multiple localities. Mol Ecol 31:4127–4145. https://doi.org/10.1111/mec.16558
- Olbertova H, Plevova K, Pavlova S, et al (2022) Evolution of TP53 abnormalities during CLL disease course is associated with telomere length changes. BMC Cancer 22:. https://doi.org/10.1186/s12885-022-09221-z
- Olinger E, eline Schaeffer C, Kidd K, et al (2022) An intermediate-effect size variant in UMOD confers risk for chronic kidney disease. https://doi.org/10.1073/pnas
- Pekár S, Dušátková LP, Macháčková T, et al (2022) Gut-content analysis in four species, combined with comparative analysis of trophic traits, suggests an araneophagous habit for the entire family Palpimanidae (Araneae). Org Divers Evol 22:265–274. https://doi.org/10.1007/s13127-021-00525-9
- Pokorna P, Giannoula ;, Klement L, et al (2022) Minimal Residual Disease-Guided Intermittent Dosing in Patients With Cancer: Successful Treatment of Chemoresistant Anaplastic Large Cell Lymphoma Using Intermittent Lorlatinib Dosing
- Pomahacova R, Paterova P, Nykodymova E, et al (2022) Cystic ovarian teratoma as a novel tumor and growth hormone deficiency as a new condition presenting in Multiple Endocrine Neoplasia type 2B: Case reports and review of the literature. Biomed Pap 166:105–111. https://doi.org/10.5507/bp.2021.051
- Rob F, Schierova D, Stehlikova Z, et al (2022) Association between ustekinumab therapy and changes in specific anti-microbial response, serum biomarkers, and microbiota composition in patients with IBD: A pilot study. PLoS One 17:e0277576. https://doi.org/10.1371/journal.pone.0277576
- Rybova J, Kuchar L, Sikora J, et al (2022) Skin inflammation and impaired adipogenesis in a mouse model of acid ceramidase deficiency. J Inherit Metab Dis 45:1175–1190. https://doi.org/10.1002/jimd.12552
- Schmiedová L, Tomášek O, Pinkasová H, et al (2022) Variation in diet composition and its relation to gut microbiota in a passerine bird. Sci Rep 12:. https://doi.org/10.1038/s41598-022-07672-9
- Schwarz M, Ryba L, Křepelová A, et al (2022) Zimmermann–Laband syndrome in monozygotic twins with a mild neurobehavioral phenotype lacking gingival overgrowth—A case report of a novel KCNN3 gene variant. Am J Med Genet Part A 188:1083–1087. https://doi.org/10.1002/ajmg.a.62616
- Sikora J, Kmochová T, Mušálková D, et al (2022) A mutation in the SAA1 promoter causes hereditary amyloid A amyloidosis. Kidney Int 101:349–359. https://doi.org/10.1016/j.kint.2021.09.007
- Slavik H, Balik V, Kokas FZ, et al (2022) Transcriptomic Profiling Revealed Lnc-GOLGA6A-1 as a Novel Prognostic Biomarker of Meningioma Recurrence. Neurosurgery 91:360–369. https://doi.org/10.1227/neu.0000000000002026
- Skalníková M, Staňo Kozubík K, Trizuljak J, et al (2022) A GP1BA Variant in a Czech Family with Monoallelic Bernard-Soulier Syndrome. Int J Mol Sci 23:. https://doi.org/10.3390/ijms23020885
- Snopková K, Dufková K, Chamrád I, et al (2022) Pyocin-mediated antagonistic interactions in Pseudomonas spp. isolated in James Ross Island, Antarctica. Environ Microbiol 24:1294–1307. https://doi.org/10.1111/1462-2920.15809
- Souche E, Beltran S, Brosens E, et al (2022) Recommendations for whole genome sequencing in diagnostics for rare diseases. Eur J Hum Genet 30:1017–1021. https://doi.org/10.1038/s41431-022-01113-x
- Souckova O, Skopova V, Baresova V, et al (2022) Metabolites of De Novo Purine Synthesis: Metabolic Regulators and Cytotoxic Compounds. Metabolites 12:. https://doi.org/10.3390/metabo12121210
- Stenton SL, Tesarova M, Sheremet NL, et al (2022) DNAJC30 defect: A frequent cause of recessive Leber hereditary optic neuropathy and Leigh syndrome. Brain 145:1624–1631. https://doi.org/10.1093/brain/awac052
- Stolfa M, Kunikova T, Novotna D, et al (2022) CASE REPORT A unique coincidence of a 17q12 deletion and duplication in a Czech family led to a refined genotype – phenotype correlation. 870–877. https://doi.org/10.1002/ajmg.a.63085
- Sporikova Z, Slavkovsky R, Tuckova L, et al (2021) IDH1/2 Mutations in Patients With Diffuse Gliomas: A Single Centre Retrospective Massively Parallel Sequencing Analysis. Appl Immunohistochem Mol Morphol. 2022 Mar 1;30(3):178-183 https://doi.org/10.1097/PAI.0000000000000997
- Vozdova M, Kubickova S, Kopecka V, et al (2022) Effects of the air pollution dynamics on semen quality and sperm DNA methylation in men living in urban industrial agglomeration. Environ Mol Mutagen 63:76–83. https://doi.org/10.1002/em.22474
Publications with the support of NCMG for the year 2021
- Brinkmann J, Lissewski C, Pinna V, et al (2021) The clinical significance of A2ML1 variants in Noonan syndrome has to be reconsidered. Eur J Hum Genet 29:524–527. https://doi.org/10.1038/s41431-020-00743-3
- Zaliova M, Winkowska L, Stuchly J, et al (2021) A novel class of ZNF384 aberrations in acute leukemia. Blood Adv 5:4393–4397. https://doi.org/10.1182/bloodadvances.2021005318
- Franková V, Driscoll RO, Jansen ME, et al (2021) Regulatory landscape of providing information on newborn screening to parents across Europe. Eur J Hum Genet 29:. https://doi.org/10.1038/s41431-020-00716-6
- Buglioni A, Hasadsri L, Nasr SH, et al (2021) Mitochondriopathy Manifesting as Inherited Tubulointerstitial Nephropathy Without Symptomatic Other Organ Involvement. Kidney Int Reports 4:2514–2518. https://doi.org/10.1016/j.ekir.2021.05.042
- Capkova Z, Capkova P, Srovnal J, et al (2021) Duplication of 9p24.3 in three unrelated patients and their phenotypes, considering affected genes, and similar recurrent variants. Mol Genet Genomic Med 9:. https://doi.org/10.1002/mgg3.1592
- Dudakova L, Stranecky V, Piherova L, et al (2021) Non-Penetrance for Ocular Phenotype in Two Individuals Carrying Heterozygous Loss-of-Function ZEB1 Alleles. Genes (Basel) 12:. https://doi.org/10.3390/genes12050677
- Lobello C, Tichy B, Bystry V, et al (2021) STAT3 and TP53 mutations associate with poor prognosis in anaplastic large cell lymphoma. Leukemia 35:. https://doi.org/10.1038/s41375-020-01093-1
- Sikora J, Kmochová T, Mušálková D, et al (2021) A mutation in the SAA1 promoter causes hereditary amyloid A amyloidosis. Kidney Int. https://doi.org/10.1016/j.kint.2021.09.007
- Klempt P, Brzoň O, Kašný M, et al (2021) Distribution of SARS-CoV-2 Lineages in the Czech Republic, Analysis of Data from the First Year of the Pandemic. Microorganisms 9:. https://doi.org/10.3390/microorganisms9081671
- Pekár S, Dušátková LP, Macháčková T, et al (2021) Gut-content analysis in four species, combined with comparative analysis of trophic traits, suggests an araneophagous habit for the entire family Palpimanidae (Araneae). Org Divers Evol. https://doi.org/10.1007/s13127-021-00525-9
- Tidu F, De Zuani M, Jose SS, et al (2021) NFAT signaling in human mesenchymal stromal cells affects extracellular matrix remodeling and antifungal immune responses. iScience 24:. https://doi.org/10.1016/j.isci.2021.102683
- Senovska A, Drozdova E, Vaculik O, et al (2021) Cost-effective straightforward method for captured whole mitogenome sequencing of ancient DNA. Forensic Sci Int 319:110638. https://doi.org/10.1016/j.forsciint.2020.110638
- Zaliova M, Potuckova E, Lukes J, et al (2021) Frequency and prognostic impact of ZEB2 H1038 and Q1072 mutations in childhood B-other acute lymphoblastic leukemia. Haematologica 106:886–890. https://doi.org/10.3324/haematol.2020.249094
- Čížková D, Ďureje Ľ, Piálek J, Kreisinger J (2021) Experimental validation of small mammal gut microbiota sampling from faeces and from the caecum after death. Heredity (Edinb) 127:141–150. https://doi.org/10.1038/s41437-021-00445-6
- Tkáčová Z, Bhide K, Mochnáčová E, et al (2021) Comprehensive Mapping of the Cell Response to Borrelia bavariensis in the Brain Microvascular Endothelial Cells in vitro Using RNA-Seq. Front Microbiol 12:1–13. https://doi.org/10.3389/fmicb.2021.760627
- Navrkalova V, Plevova K, Hynst J, et al (2021) LYmphoid NeXt-Generation Sequencing (LYNX) Panel: A Comprehensive Capture-Based Sequencing Tool for the Analysis of Prognostic and Predictive Markers in Lymphoid Malignancies. J Mol Diagnostics 23:959–974. https://doi.org/10.1016/j.jmoldx.2021.05.007
- Bencurova P, Baloun J, Hynst J, et al (2021) Dynamic miRNA changes during the process of epileptogenesis in an infantile and adult-onset model. Sci Rep 11:1–14. https://doi.org/10.1038/s41598-021-89084-9
- Snopková K, Dufková K, Chamrád I, et al (2021) Pyocin‐mediated antagonistic interactions in Pseudomonas spp. isolated in James Ross Island, Antarctica. Environ Microbiol. https://doi.org/10.1111/1462-2920.15809
- Hynst J, Navrkalova V, Pal K, Pospisilova S (2021) Bioinformatic strategies for the analysis of genomic aberrations detected by targeted NGS panels with clinical application. PeerJ 9:e10897. https://doi.org/10.7717/peerj.10897
- KISS D, MACHACKOVA T, SOUCKOVA K, et al (2021) An independent validation study of candidate micrornas as predictive biomarkers for bevacizumab-based therapy in patients with metastatic colorectal cancer. In Vivo (Brooklyn) 35:2809–2814. https://doi.org/10.21873/INVIVO.12567
- Jiménez-Munguía I, Tomečková Z, Mochnáčová E, et al (2021) Transcriptomic analysis of human brain microvascular endothelial cells exposed to laminin binding protein (adhesion lipoprotein) and Streptococcus pneumoniae. Sci Rep 11:1–15. https://doi.org/10.1038/s41598-021-87021-4
- Sedlar K, Vasylkivska M, Musilova J, et al (2021) Phenotypic and genomic analysis of isopropanol and 1,3-propanediol producer Clostridium diolis DSM 15410. Genomics 113:1109–1119. https://doi.org/10.1016/j.ygeno.2020.11.007
- Srovnal J, Vidlarova M, Berta E, et al (2021) Abstract 665: Cannabinoid receptor 2 tumor tissue gene expression positively affects lung cancer survival following radical surgery. In: Clinical Research (Excluding Clinical Trials). American Association for Cancer Research, pp 665–665
- Malcikova J, Pavlova S, Kunt Vonkova B, et al (2021) Low-burden TP53 mutations in CLL: Clinical impact and clonal evolution within the context of different treatment options. Blood. https://doi.org/10.1182/blood.2020009530
- Schierova D, Roubalova R, Kolar M, et al (2021) Fecal Microbiome Changes and Specific Anti-Bacterial Response in Patients with IBD during Anti-TNF Therapy. Cells 10:3188. https://doi.org/10.3390/cells10113188
- Perestrelo AR, Hejret V, Varadarajan NM, et al (2021) The mechanical regulation of RNA binding protein hnRNPC in the failing heart hnRNPC is a mechanosensitive component of RNA homeostasis apparatus in heart failure . Giancarlo Forte , PhD Center for Translational Medicine ( CTM ) International Clinical Rese. BioarxivRxiv 1–44. https://doi.org/10.1101/2021.08.27.457906
- Sharma S, Pavlasova GM, Seda V, et al (2021) miR-29 modulates CD40 signaling in chronic lymphocytic leukemia by targeting TRAF4: an axis affected by BCR inhibitors. Blood 137:2481–2494. https://doi.org/10.1182/blood.2020005627
- Kripnerová M, Parmar HS, Šána J, et al (2021) Complex interplay of genes underlies invasiveness in fibrosarcoma progression model. J Clin Med 10:2297. https://doi.org/10.3390/jcm10112297
- Xiao W, Ren L, Chen Z, et al (2021) Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat Biotechnol 39:1141–1150. https://doi.org/10.1038/s41587-021-00994-5
- Burska D, Stiburek L, Krizova J, et al (2021) Homozygous missense mutation in UQCRC2 associated with severe encephalomyopathy, mitochondrial complex III assembly defect and activation of mitochondrial protein quality control. Biochim Biophys Acta - Mol Basis Dis 1867:166147. https://doi.org/10.1016/j.bbadis.2021.166147
- Sottas C, Schmiedová L, Kreisinger J, et al (2021) Gut microbiota in two recently diverged passerine species: evaluating the effects of species identity, habitat use and geographic distance. BMC Ecol Evol 21:1–14. https://doi.org/10.1186/s12862-021-01773-1
- Fajkus P, Kilar A, Nelson ADL, et al (2021) Evolution of plant telomerase RNAs: Farther to the past, deeper to the roots. Nucleic Acids Res 49:7680–7694. https://doi.org/10.1093/nar/gkab545
- Bleyer AJ, Wolf MT, Kidd KO, et al (2021) Autosomal dominant tubulointerstitial kidney disease: more than just HNF1β. Pediatr. Nephrol.
- Bene Z, Fejes Z, Szanto TG, et al (2021) Enhanced Expression of Human Epididymis Protein 4 (HE4) Reflecting Pro-Inflammatory Status Is Regulated by CFTR in Cystic Fibrosis Bronchial Epithelial Cells. Front Pharmacol 12:1–16. https://doi.org/10.3389/fphar.2021.592184
- Yiallouros PK, Matthaiou A, Anagnostopoulou P, et al (2021) Demographic characteristics, clinical and laboratory features, and the distribution of pathogenic variants in the CFTR gene in the Cypriot cystic fibrosis (CF) population demonstrate the utility of a national CF patient registry. Orphanet J Rare Dis 16:1–21. https://doi.org/10.1186/s13023-021-02049-z
- Oláhová M, Peter B, Szilagyi Z, et al (2021) POLRMT mutations impair mitochondrial transcription causing neurological disease. Nat Commun 12:1–13. https://doi.org/10.1038/s41467-021-21279-0
- Fišarová L, Botka T, Du X, et al (2021) Staphylococcus epidermidis Phages Transduce Antimicrobial Resistance Plasmids and Mobilize Chromosomal Islands. mSphere 6:1–19. https://doi.org/10.1128/mSphere.00223-21
- Bartos M, Siegl F, Kopkova A, et al (2021) Small RNA Sequencing Identifies PIWI-Interacting RNAs Deregulated in Glioblastoma—piR-9491 and piR-12488 Reduce Tumor Cell Colonies In Vitro. Front Oncol 11:1–11. https://doi.org/10.3389/fonc.2021.707017
- Lahrouchi N, Postma A V., Salazar CM, et al (2021) Biallelic loss-of-function variants in PLD1 cause congenital right-sided cardiac valve defects and neonatal cardiomyopathy. J Clin Invest 131:1–12. https://doi.org/10.1172/JCI142148
- Olinger E, Schaeffer C, Kidd K, et al (2021) An intermediate effect size variant in UMOD confers risk for chronic kidney disease. medRxiv 1–28. https://doi.org/https://doi.org/10.1101/2021.09.27.21263789
- Dyková I, Žák J, Reichard M, et al (2021) Histopathology of laboratory-reared Nothobranchius fishes: Mycobacterial infections versus neoplastic lesions. J Fish Dis 44:1179–1190. https://doi.org/10.1111/jfd.13378
- Jeanne M, Demory H, Moutal A, et al (2021) Missense variants in DPYSL5 cause a neurodevelopmental disorder with corpus callosum agenesis and cerebellar abnormalities. Am J Hum Genet 108:951–961. https://doi.org/10.1016/j.ajhg.2021.04.004
- Liang HC, Costanza M, Prutsch N, et al (2021) Super-enhancer-based identification of a BATF3/IL-2R−module reveals vulnerabilities in anaplastic large cell lymphoma. Nat. Commun. 12
- Sági JC, Gézsi A, Egyed B, et al (2021) Pharmacogenetics of the central nervous system—toxicity and relapse affecting the cns in pediatric acute lymphoblastic leukemia. Cancers (Basel) 13:. https://doi.org/10.3390/cancers13102333
- Chowdhury F, Wang L, Al-Raqad M, et al (2021) Haploinsufficiency of PRR12 causes a spectrum of neurodevelopmental, eye, and multisystem abnormalities. Genet Med 23:1234–1245. https://doi.org/10.1038/s41436-021-01129-6
- Parimuchová A, Dušátková LP, Kováč Ľ, et al (2021) The food web in a subterranean ecosystem is driven by intraguild predation. Sci Rep 11:1–11. https://doi.org/10.1038/s41598-021-84521-1
- Vylet’Al P, Kidd K, Ainsworth HC, et al (2021) Plasma Mucin-1 (CA15-3) Levels in Autosomal Dominant Tubulointerstitial Kidney Disease due to MUC1 Mutations. Am J Nephrol 52:378–387. https://doi.org/10.1159/000515810
- Kalita O, Sporikova Z, Hajduch M, et al (2021) The influence of gene aberrations on survival in resected idh wildtype glioblastoma patients: A single-institution study. Curr Oncol 28:1280–1293. https://doi.org/10.3390/curroncol28020122
Publications with the support of NCMG for the year 2020
- Skalicka P, Porter LF, Brejchova K, et al (2020) Brittle cornea syndrome: A systemic review of disease-causing mutations in ZNF469 and two novel variants identified in a patient followed for 26 years. Biomed Pap. Biomed Pap 164:183–188. doi: 10.5507/bp.2019.017
- Majer F, Kousal B, Dusek P, et al (2020) Alu-mediated Xq24 deletion encompassing CUL4B, LAMP2, ATP1B4, TMEM255A, and ZBTB33 genes causes Danon disease in a female patient. Am J Med Genet Part A 182:219–223. doi: 10.1002/ajmg.a.61416
- Kubánek M, Schimerová T, Piherová L, et al (2020) Desminopathy: Novel Desmin Variants, a New Cardiac Phenotype, and Further Evidence for Secondary Mitochondrial Dysfunction. J Clin Med 9:937. doi: 10.3390/jcm9040937
- Larose H, Prokoph N, Matthews JD, et al (2020) Whole Exome Sequencing reveals NOTCH1 mutations in anaplastic large cell lymphoma and points to Notch both as a key pathway and a potential therapeutic target. Haematologica 105:haematol.2019.238766. doi: 10.3324/haematol.2019.238766
- Jedličková I, Cadieux-Dion M, Přistoupilová A, et al (2020) Autosomal-dominant adult neuronal ceroid lipofuscinosis caused by duplication in DNAJC5 initially missed by Sanger and whole-exome sequencing. Eur J Hum Genet 87:579–84. doi: 10.1038/s41431-019-0567-2
- Kozlova V, Ledererova A, Ladungova A, et al (2020) CD20 is dispensable for B-cell receptor signaling but is required for proper actin polymerization, adhesion and migration of malignant B cells. PLoS One 15:1–20. doi: 10.1371/journal.pone.0229170
- Vrbovská V, Sedláček I, Zeman M, et al (2020) Characterization of staphylococcus intermedius group isolates associated with animals from antarctica and emended description of staphylococcus delphini. Microorganisms 8:1–18. doi: 10.3390/microorganisms8020204
- Bene Z, Fejes Z, Macek M, et al (2020) Laboratory biomarkers for lung disease severity and progression in cystic fibrosis. Clin Chim Acta. doi: 10.1016/j.cca.2020.05.015
- Cormican S, Kennedy C, Connaughton DM, et al (2020) Renal transplant outcomes in patients with autosomal dominant tubulointerstitial kidney disease. Clin Transplant 34:1–6. doi: 10.1111/ctr.13783
- Vozdova M, Kubickova S, Pal K, et al (2020) Recurrent gene mutations detected in canine mast cell tumours by next generation sequencing. Vet Comp Oncol 1–10. doi: 10.1111/vco.12572
- Sottas C, Reif J, Kreisinger J, et al (2020) Tracing the early steps of competition-driven eco-morphological divergence in two sister species of passerines. Evol. Ecol.
- Mazurova S, Tesarova M, Zeman J, et al (2020) Fatal neonatal nephrocutaneous syndrome in 18 Roma children with EGFR deficiency. J Dermatol 1–6. doi: 10.1111/1346-8138.15317
- Leeksma AC, Baliakas P, Moysiadis T, et al (2020) Genomic arrays identify high-risk chronic lymphocytic leukemia with genomic complexity: a multi-center study. Haematologica haematol.2019.239947. doi: 10.3324/haematol.2019.239947
- Olinger E, Hofmann P, Kidd K, et al (2020) Clinical and Genetic Spectra of Autosomal Dominant Tubulointerstitial Kidney Disease due to Mutations in UMOD and MUC1. Kidney Int. doi: 10.1016/j.kint.2020.04.038
- Lhotova K, Stolarova L, Zemankova P, et al (2020) Multigene panel germline testing of 1333 Czech patients with ovarian cancer. Cancers (Basel) 12:3–16. doi: 10.3390/cancers12040956
- Peska V, Mand T, Vitales D, et al (2020) Human-like telomeres in Zostera marina reveal a mode of transition from the plant to the human telomeric sequences. bioRxiv. doi: doi.org/10.1101/2020.03.11.987156
- Slavik H, Balik V, Vrbkova J, et al (2020) Identification of Meningioma Patients at High Risk ofTumor Recurrence Using MicroRNA Profiling. Neurosurgery 0:1–9. doi: 10.1093/neuros/nyaa009
- Sutton L-A, Ljungström V, et al (2020) Comparative analysis of targeted next-generation sequencing panels for the detection of gene mutations in chronic lymphocytic leukemia: an ERIC multi-center study. Haematologica 105:haematol.2019.234716. doi: 10.3324/haematol.2019.234716
- Tkáčiková Ľ, Mochnáčová E, Tyagi P, et al (2020) Comprehensive mapping of the cell response to E. coli infection in porcine intestinal epithelial cells pretreated with exopolysaccharide derived from Lactobacillus reuteri. Vet Res 51:1–13. doi: 10.1186/s13567-020-00773-1
- Snopková K, Čejková D, Dufková K, et al (2020) Genome sequences of two Antarctic strains of Pseudomonas prosekii: insights into adaptation to extreme conditions. Arch Microbiol 202:447–454. doi: 10.1007/s00203-019-01755-4Arch Microbiol. doi: 10.1007/s00203-019-01755-4
- Bleyer AJ, Kidd K, Robins V, et al (2020) Outcomes of patient self-referral for the diagnosis of several rare inherited kidney diseases. Genet Med 22:142–149. doi: 10.1038/s41436-019-0617-8Genet Med 0:1–8. doi: 10.1038/s41436-019-0617-8
- Lukes J, Danek P, Alejo-Valle O, et al (2020) Chromosome 21 gain is dispensable for transient myeloproliferative disorder driven by a novel GATA1 mutation. Leukemia. doi: 10.1038/s41375-020-0769-1
- Laidou S, Alanis-Lobato G, Pribyl J, et al (2020) Nuclear inclusions of pathogenic ataxin-1 induce oxidative stress and perturb the protein synthesis machinery. Redox Biol 32:101458. doi: 10.1016/j.redox.2020.101458
- Dudakova L, Skalicka P, Ulmanová O, et al (2020) Pseudodominant Nanophthalmos in a Roma Family Caused by a Novel PRSS56 Variant. J Ophthalmol 2020:1–9. doi: 10.1155/2020/6807809
- Jedličková I, Přistoupilová A, Nosková L, et al (2020) Spinal muscular atrophy caused by a novel Alu-mediated deletion of exons 2a-5 in SMN1 undetectable with routine genetic testing. Mol Genet genomic Med e1238. doi: 10.1002/mgg3.1238
- Geryk J, Krsička D, Vlčková M, et al (2020) The Key Role of Purine Metabolism in the Folate-Dependent Phenotype of Autism Spectrum Disorders: An In Silico Analysis. Metabolites 10:. doi: 10.3390/metabo10050184
- Schizophrenia Working Group of the Psychiatric Genomics Consortium., Ripke S, Walters JT, O’Donovan MC (2020) Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv 2020.09.12.20192922
- Pěnčík O, Jiráčková K, Mašlaňová I, et al (2020) Spatial structure of permafrost and active layer from the James Ross Island , Antarctica : Geological and Microbiological Insights. CZECH POLAR REPORTS 10:127–128
- Jedlickova I, Pristoupilova A, Hulkova H, et al (2020) NOTCH2NLC CGG Repeats Are Not Expanded and Skin Biopsy Was Negative in an Infantile Patient With Neuronal Intranuclear Inclusion Disease. J Neuropathol Exp Neurol 79:1065–1071. https://doi.org/10.1093/jnen/nlaa070
- Stolarova L, Jelinkova S, Storchova R, et al (2020) Identification of Germline Mutations in Melanoma Patients with Early Onset, Double Primary Tumors, or Family Cancer History by NGS Analysis of 217 Genes. Biomedicines 8:404. https://doi.org/10.3390/biomedicines8100404
- Živná M, Kidd K, Zaidan M, et al (2020) An International Cohort Study of Autosomal Dominant Tubulointerstitial Kidney Disease due to REN Mutations Identifies Distinct Clinical Subtypes. Kidney Int. https://doi.org/10.1016/j.kint.2020.06.041
- Schneeberger PE, Kortüm F, Korenke GC, et al (2020) Biallelic MADD variants cause a phenotypic spectrum ranging from developmental delay to a multisystem disorder. Brain 143:2437–2453. https://doi.org/10.1093/brain/awaa204
- Franková V, Driscoll RO, Jansen ME, et al (2020) Regulatory landscape of providing information on newborn screening to parents across Europe. Eur J Hum Genet. https://doi.org/10.1038/s41431-020-00716-6
- Brinkmann J, Lissewski C, Pinna V, et al (2020) The clinical significance of A2ML1 variants in Noonan syndrome has to be reconsidered. Eur J Hum Genet 3–6. https://doi.org/10.1038/s41431-020-00743-3
Publications with the support of NCMG for the year 2019
- Hernández-Sánchez M, Kotaskova J, Rodríguez AE, et al (2019) CLL cells cumulate genetic aberrations prior to the first therapy even in outwardly inactive disease phase. Leukemia 33:518–558. doi: 10.1038/s41375-018-0255-1
- Cormican S, Connaughton DM, Kennedy C, et al (2019) Autosomal dominant tubulointerstitial kidney disease (ADTKD) in Ireland. Ren Fail 41:832–841. doi: 10.1080/0886022x.2019.1655452
- Bereshchenko O, Lo Re O, Nikulenkov F, et al (2019) Deficiency and haploinsufficiency of histone macroH2A1.1 in mice recapitulate hematopoietic defects of human myelodysplastic syndrome. Clin Epigenetics 11:1–14. doi: 10.1186/s13148-019-0724-z
- Balik V, Takizawa K (2019) Safe and bloodless exposure of the third segment of the vertebral artery: a step-by-step overview based on over 50 personal cases. Neurosurg Rev. doi: 10.1007/s10143-019-01158-5
- Fajkus P, Peška V, Závodník M, et al (2019) Telomerase RNAs in land plants. Nucleic Acids Res 1–15. doi: 10.1093/nar/gkz695
- Brejchova K, Dudakova L, Skalicka P, et al (2019) IPSC-Derived Corneal Endothelial-like Cells Act as an Appropriate Model System to Assess the Impact of SLC4A11 Variants on Pre-mRNA Splicing. Investig Opthalmology Vis Sci 60:3084. doi: 10.1167/iovs.19-26930
- Petrova G, Yaneva N, Hrbková J, et al (2019) Identification of 99% of CFTR gene mutations in Bulgarian‐, Bulgarian Turk‐, and Roma cystic fibrosis patients. Mol. Genet. Genomic Med. 7
- Souček P, Réblová K, Kramárek M, et al (2019) High-throughput analysis revealed mutations’ diverging effects on SMN1 exon 7 splicing. RNA Biol 16:1364–1376. doi: 10.1080/15476286.2019.1630796
- Kostovcikova K, Coufal S, Galanova N, et al (2019) Diet rich in animal protein promotes pro-inflammatory macrophage response and exacerbates colitis in mice. Front Immunol 10:1–14. doi: 10.3389/fimmu.2019.00919
- Oliver-De La Cruz J, Nardone G, Vrbsky J, et al (2019) Substrate mechanics controls adipogenesis through YAP phosphorylation by dictating cell spreading. Biomaterials 205:64–80. doi: 10.1016/j.biomaterials.2019.03.009
- Jelinkova S, Fojtik P, Kohutova A, et al (2019) Dystrophin Deficiency Leads to Genomic Instability in Human Pluripotent Stem Cells via NO Synthase-Induced Oxidative Stress. Cells 8:53. doi: 10.3390/cells8010053
- Mareckova A, Malcikova J, Tom N, et al (2018) ATM and TP53 mutations show mutual exclusivity but distinct clinical impact in mantle cell lymphoma patients. Leuk Lymphoma 60:1420–1428. doi: 10.1080/10428194.2018.1542144
- Králová S, Busse HJ, Švec P, et al (2019) Flavobacterium circumlabens sp. nov. and Flavobacterium cupreum sp. nov., two psychrotrophic species isolated from Antarctic environmental samples. Syst Appl Microbiol 42:291–301. doi: 10.1016/j.syapm.2018.12.005
- Kleiblová P, Stolařová L, Křížová K, et al (2019) Germline CHEK2 Gene Mutations in Hereditary Breast Cancer Predisposition – Mutation Types and their Biological and Clinical Relevance. Klin Onkol 32:36–50. doi: 10.14735/amko2019s36
- Bleyer AJ, Kmoch S, Greka A (2019) Diagnostic Utility of Exome Sequencing for Kidney Disease. N Engl J Med 380:2080. doi: 10.1056/NEJMc1903250
- Wayhelova M, Oppelt J, Smetana J, et al (2019) Novel de novo frameshift variant in the ASXL3 gene in a child with microcephaly and global developmental delay.Mol Med Rep 20:505–512. doi: 10.3892/mmr.2019.10303
- Doubková M, Staňo Kozubík K, Radová L, et al (2019) A novel germline mutation of the SFTPA1 gene in familial interstitial pneumonia. Hum Genome Var 6:12. doi: 10.1038/s41439-019-0044-z
- Zaliova M, Stuchly J, Winkowska L, et al (2019) Genomic landscape of pediatric B-other acute lymphoblastic leukemia in a consecutive European cohort. Haematologica 104:1396–1406. doi: 10.3324/haematol.2018.204974
- Nagy B, Bene Z, Fejes Z, et al (2019) Human epididymis protein 4 (HE4) levels inversely correlate with lung function improvement (delta FEV 1 ) in cystic fibrosis patients receiving ivacaftor treatment. J Cyst Fibros 18:271–277. doi: 10.1016/j.jcf.2018.08.013
- Zaliova M, Potuckova E, Hovorkova L, et al (2019) ERG deletions in childhood acute lymphoblastic leukemia with DUX4 rearrangements are mostly polyclonal, prognostically relevant and their detection rate strongly depends on method’s sensitivity. Haematologica 104:1407–1416. doi: 10.3324/haematol.2018.204487
- Wiatrek DM, Candela ME, Sedmík J, et al (2019) Activation of innate immunity by mitochondrial dsRNA in mouse cells lacking p53 protein. Rna 25:713–726. doi: 10.1261/rna.069625.118
- Kleiblova P, Stolarova L, Krizova K, et al (2019) Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int J Cancer 145:1782–1797. doi: 10.1002/ijc.32385
- Holub D, Flodrova P, Pika T, et al (2019) Mass Spectrometry Amyloid Typing Is Reproducible across Multiple Organ Sites. Biomed Res Int 2019:1–9. doi: 10.1155/2019/3689091
- Dudakova L, Evans CJ, Pontikos N, et al (2019) The utility of massively parallel sequencing for posterior polymorphous corneal dystrophy type 3 molecular diagnosis. Exp Eye Res 182:160–166. doi: 10.1016/j.exer.2019.03.002
- Vevera J, Zarrei M, Hartmannová H, et al (2019) Rare copy number variation in extremely impulsively violent males.Genes, Brain Behav 18:e12536. doi: 10.1111/gbb.12536
- Pavlova S, Smardova J, Tom N, Trbusek M (2019) Detection and Functional Analysis of TP53 Mutations in CLL. In: Methods in molecular biology (Clifton, N.J.). pp 63–81
- Yu FP, Sajdak BS, Sikora J, et al (2019) Acid Ceramidase Deficiency in Mice Leads to Severe Ocular Pathology and Visual Impairment. Am J Pathol 189:320–338. doi: 10.1016/j.ajpath.2018.10.018
- Yu FPS, Molino S, Sikora J, et al (2019) Hepatic pathology and altered gene transcription in a murine model of acid ceramidase deficiency. Lab Investig 99:1572–1592. doi: 10.1038/s41374-019-0271-4
- Devuyst O, Olinger E, Weber S, et al (2019) Autosomal dominant tubulointerstitial kidney disease. Nat Rev Dis Prim 5:60. doi: 10.1038/s41572-019-0109-9
- Nunvar J, Hogan AM, Buroni S, et al (2019) The Effect of 2-Thiocyanatopyridine Derivative 11026103 on Burkholderia Cenocepacia: Resistance Mechanisms and Systemic Impact. Antibiotics 8:159. doi: 10.3390/antibiotics8040159
- Machálková M, Pavlatovská B, Michálek J, et al (2019) Drug Penetration Analysis in 3D Cell Cultures Using Fiducial-Based Semiautomatic Coregistration of MALDI MSI and Immunofluorescence Images. Anal Chem 91:13475–13484. doi: 10.1021/acs.analchem.9b02462
- Pelet A, Skopova V, Steuerwald U, et al (2019) PAICS deficiency, a new defect of de novo purine synthesis resulting in multiple congenital anomalies and fatal outcome. Hum Mol Genet 1–30. doi: 10.1093/hmg/ddz237
- Sedlar K, Kolek J, Gruber M, et al (2019) A transcriptional response of Clostridium beijerinckii NRRL B-598 to a butanol shock. Biotechnol Biofuels 12:243. doi: 10.1186/s13068-019-1584-7
- Káňová E, Tkáčová Z, Bhide K, et al (2019) Transcriptome analysis of human brain microvascular endothelial cells response to Neisseria meningitidis and its antigen MafA using RNA-seq. Sci Rep 9:18763. doi: 10.1038/s41598-019-55409-y
- Capkova Z, Capkova P, Srovnal J, et al (2019) Differences in the importance of microcephaly, dysmorphism, and epilepsy in the detection of pathogenic CNVs in ID and ASD patients.PeerJ 2019:e7979. doi: 10.7717/peerj.7979
- Stehlikova Z, Tlaskal V, Galanova N, et al (2019) Oral Microbiota Composition and Antimicrobial Antibody Response in Patients with Recurrent Aphthous Stomatitis. Microorganisms 7:636. doi: 10.3390/microorganisms7120636
- Vasylkivska M, Jureckova K, Branska B, et al (2019) Transcriptional analysis of amino acid, metal ion, vitamin and carbohydrate uptake in butanol-producing Clostridium beijerinckii NRRL B-598. PLoS One 14:e0224560. doi: 10.1371/journal.pone.0224560
- Bleyer AJ, Kidd K, Johnson E, et al (2019) Quality of life in patients with autosomal dominant tubulointerstitial kidney disease. Clin Nephrol 92:302–311. doi: 10.5414/CN109842
Publications with the support of NCMG for the year 2018
- Gstrein T, Edwards A, Přistoupilová A, et al (2018) Mutations in Vps15 perturb neuronal migration in mice and are associated with neurodevelopmental disease in humans. Nat Neurosci 1. doi: 10.1038/s41593-017-0053-5
- Dudakova L, Vercruyssen JHJ, Balikova I, et al (2018) Analysis of KERA in four families with cornea plana identifies two novel mutations. Acta Ophthalmol 96:e87–e91. doi: 10.1111/aos.13484
- Dudakova L, Cheong S-S, Merjava SR, et al (2018) Familial Limbal Stem Cell Deficiency: Clinical, Cytological and Genetic Characterization. Stem Cell Rev Reports 14:148–151. doi: 10.1007/s12015-017-9780-y
- Paderova J, Drabova J, Holubova A, et al (2018) Under the mask of Kabuki syndrome: Elucidation of genetic-and phenotypic heterogeneity in patients with Kabuki-like phenotype. Eur J Med Genet. doi: 10.1016/j.ejmg.2018.01.00
- van den Ameele J, Jedlickova I, Pristoupilova A, et al (2018) Teenage-onset progressive myoclonic epilepsy due to a familial C9orf72 repeat expansion. Neurology doi: 10.1212/WNL.0000000000004999
- Brazdilova K, Plevova K, Skuhrova Francova H, et al (2018) Multiple productive IGH rearrangements denote oligoclonality even in immunophenotypically monoclonal CLL. Leukemia 32:234–236. doi: 10.1038/leu.2017.274
- Kremlikova Pourova R, Paderova J, Copikova J, et al (2018) SD-OCT imaging as a valuable tool to support molecular genetic diagnostics of Usher syndrome type 1. J Am Assoc Pediatr Ophthalmol Strabismus 1–3. doi: 10.1016/j.jaapos.2017.12.009
- Liskova P, Dudakova L, Evans CJ, et al (2018) Ectopic GRHL2 Expression Due to Non-coding Mutations Promotes Cell State Transition and Causes Posterior Polymorphous Corneal Dystrophy 4. Am J Hum Genet 102:447–459. doi: 10.1016/j.ajhg.2018.02.002
- Zikánová M, Wahezi D, Hay A, et al (2018) Clinical manifestations and molecular aspects of phosphoribosylpyrophosphate synthetase superactivity in females. Rheumatology 1–6. doi: 10.1093/rheumatology/key041
- Slamova Z, Nazaryan-Petersen L, Mehrjouy MM, et al (2018) Very short DNA segments can be detected and handled by the repair machinery during germline chromothriptic chromosome reassembly. Hum Mutat. doi: 10.1002/humu.23408
- Springer D, Loucky J, Tesner P, et al (2018) Importance of the integrated test in the Down’s syndrome screening algorithm. J Med Screen 096914131775253. doi: 10.1177/0969141317752533
- Ivády G, Madar L, Dzsudzsák E, et al (2018) Analytical parameters and validation of homopolymer detection in a pyrosequencing-based next generation sequencing system. BMC Genomics 19:1–8. doi: 10.1186/s12864-018-4544-x
- Pafčo B, Čížková D, Kreisinger J, et al (2018) Metabarcoding analysis of strongylid nematode diversity in two sympatric primate species. Sci Rep 8:5933. doi: 10.1038/s41598-018-24126-3
- Malcikova J, Tausch E, Rossi D, et al (2018) ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-update on methodological approaches and results interpretation. Leukemia 1–11. doi: 10.1038/s41375-017-0007-7
- Vlčková K, Kreisinger J, Pafčo B, et al (2018) Diversity of Entamoeba spp. in African great apes and humans: an insight from Illumina MiSeq high-throughput sequencing. Int J Parasitol. doi: 10.1016/j.ijpara.2017.11.008
- Sedlar K, Koscova P, Vasylkivska M, et al (2018) Transcription profiling of butanol producer Clostridium beijerinckii NRRL B-598 using RNA-Seq. BMC Genomics 19:415. doi: 10.1186/s12864-018-4805-8
- Kutilova I, Janecko N, Cejkova D, et al (2018) Characterization of blaKPC-3-positive plasmids from an Enterobacter aerogenes isolated from a corvid in Canada. J Antimicrob Chemother. doi: 10.1093/jac/dky199
- Čížková D, Baird SJE, Těšíková J, et al (2018) Host subspecific viral strains in European house mice: Murine cytomegalovirus in the Eastern ( Mus musculus musculus ) and Western house mouse ( Mus musculus domesticus ). Virology 521:92–98. doi: 10.1016/j.virol.2018.05.023
- Tom N, Tom O, Malcikova J, et al (2018) ToTem: a tool for variant calling pipeline optimization. BMC Bioinformatics 19:243. doi: 10.1186/s12859-018-2227-x
- Evans CJ, Dudakova L, Skalicka P, et al (2018) Schnyder corneal dystrophy and associated phenotypes caused by novel and recurrent mutations in the UBIAD1 gene. BMC Ophthalmol 18:250. doi: 10.1186/s12886-018-0918-8
- Majer F, Piherova L, Reboun M, et al (2018) LAMP2 exon-copy number variations in Danon disease heterozygote female probands: Infrequent or underdetected? Am J Med Genet Part A. doi: 10.1002/ajmg.a.40430
- Agrawal K, Das V, Táborská N, et al (2018) Differential Regulation of Methylation-Regulating Enzymes by Senescent Stromal Cells Drives Colorectal Cancer Cell Response to DNA-Demethylating Epi-Drugs. Stem Cells Int 2018:1–11. doi: 10.1155/2018/6013728
- Farrell P, Férec C, Macek M, et al (2018) Estimating the age of p.(Phe508del) with family studies of geographically distinct European populations and the early spread of cystic fibrosis. Eur J Hum Genet 1. doi: 10.1038/s41431-018-0234-z
- Hiatt SM, Neu MB, Ramaker RC, et al (2018) De novo mutations in the GTP/GDP-binding region of RALA, a RAS-like small GTPase, cause intellectual disability and developmental delay. PLOS Genet 14:e1007671. doi: 10.1371/journal.pgen.1007671
- Živná M, Kidd K, Přistoupilová A, et al (2018) Noninvasive Immunohistochemical Diagnosis and Novel MUC1 Mutations Causing Autosomal Dominant Tubulointerstitial Kidney Disease. J Am Soc Nephrol 29:2418–2431. doi: 10.1681/ASN.2018020180
- Trizuljak J, Kozubík KS, Radová L, et al (2018) A novel germline mutation in GP1BA gene N-terminal domain in monoallelic Bernard-Soulier syndrome. Platelets 1–7. doi: 10.1080/09537104.2018.1529300
- Gast C, Marinaki A, Arenas-Hernandez M, et al (2018) Autosomal dominant tubulointerstitial kidney disease-UMOD is the most frequent non polycystic genetic kidney disease. BMC Nephrol 19:301. doi: 10.1186/s12882-018-1107-y
- Čaplovičová M, Moslerová V, Dupej J, et al (2018) Modeling age-specific facial development in Williams-Beuren-, Noonan-, and 22q11.2 deletion syndromes in cohorts of Czech patients aged 3-18 years: A cross-sectional three-dimensional geometric morphometry analysis of their facial gestalt. Am J Med Genet Part A. doi: 10.1002/ajmg.a.40659
- Soukupova J, Zemankova P, Lhotova K, et al (2018) Validation of CZECANCA (CZEch CAncer paNel for Clinical Application) for targeted NGS-based analysis of hereditary cancer syndromes. PLoS One 13:e0195761. doi: 10.1371/journal.pone.0195761
- Puchmajerová A, Tornikidis J, Mrňa L, et al (2018) Hereditární formy karcinomu prsu : genetická etiologie a současné možnosti prevence a chirurgické léčby. Čas Lék čes 157:90–95
- Hlavac V, Kovacova M, Elsnerova K, et al (2018) Use of Germline Genetic Variability for Prediction of Chemoresistance and Prognosis of Breast Cancer Patients. Cancers (Basel) 10:511. doi: 10.3390/cancers10120511
- Kmoch S, Zeman J (2018) Moderní metody v diagnostice a výzkumu genetických příčin vzácných onemocnění. Cas Lek Cesk 157:133–136
- Harper JC, Aittomäki K, Borry P, et al (2018) Recent developments in genetics and medically-assisted reproduction: from research to clinical applications. Hum Reprod Open. doi: 10.1093/hropen/hox015
- Yu FP, Islam D, Sikora J, et al (2018) Chronic lung injury and impaired pulmonary function in a mouse model of acid ceramidase deficiency. Am J Physiol Cell Mol Physiol ajplung.00223.2. doi: 10.1152/ajplung.00223.2017
Publications with the support of NCMG for the year 2017
- Pal K, Bystry V, Reigl T, et al (2017) GLASS: assisted and standardized assessment of gene variations from Sanger sequence trace data. Bioinformatics 33:3802–3804. doi: 10.1093/bioinformatics/btx423
- Kim Y, Park S-J, Manson SR, et al (2017) Elevated urinary CRELD2 is associated with endoplasmic reticulum stress-mediated kidney disease. JCI insight. doi: 10.1172/jci.insight.92896
- Kristoffersson U, Macek M (2017) From Mendel to Medical Genetics. Eur J Hum Genet 25:S53–S58. doi: 10.1038/ejhg.2017.157
- Hojny J, Zemankova P, Lhota F, et al (2017) Multiplex PCR and NGS-based identification of mRNA splicing variants: Analysis of BRCA1 splicing pattern as a model. Gene 637:41–49. doi: 10.1016/j.gene.2017.09.025
- Bencurova P, Baloun J, Musilova K, et al (2017) MicroRNA and mesial temporal lobe epilepsy with hippocampal sclerosis: Whole miRNome profiling of human hippocampus. Epilepsia 58:1782–1793. doi: 10.1111/epi.13870
- Dudakova L, Stranecky V, Ulmanova O, et al (2017) Segregation of a novel p.(Ser270Tyr) MAF mutation and p.(Tyr56∗) CRYGD variant in a family with dominantly inherited congenital cataracts. Mol Biol Rep. doi: 10.1007/s11033-017-4121-4
- Růžička K, Zhang M, Campilho A, et al (2017) Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol 215:157–172. doi: 10.1111/nph.14586
- Krsička D, Geryk J, Vlčková M, et al (2017) Identification of likely associations between cerebral folate deficiency and complex genetic- and metabolic pathogenesis of autism spectrum disorders by utilization of a pilot interaction modeling approach. Autism Res. doi: 10.1002/aur.1780
- Liskova P, Dudakova L, Krepelova A, et al (2017) Replication of SNP associations with keratoconus in a Czech cohort. PLoS One 12:e0172365. doi: 10.1371/journal.pone.0172365
- Mazurova S, Magner M, Kucerova-Vidrova V, et al (2017) Thymidine kinase 2 and alanyl-tRNA synthetase 2 deficiencies cause lethal mitochondrial cardiomyopathy: case reports and review of the literature. Cardiol Young 27:936–944. doi: 10.1017/S1047951116001876
Publications with the support of NCMG for the year 2016
- Nagy B, Nagy B, Fila L, et al (2016) Human Epididymis Protein 4: A Novel Serum Inflammatory Biomarker in Cystic Fibrosis. Chest 150:661–72. doi: 10.1016/j.chest.2016.04.006
- Paděrová J, Holubová A, Simandlová M, et al (2016) Molecular genetic analysis in 14 Czech Kabuki syndrome patients is confirming the utility of phenotypic scoring. Clin Genet 90:230–7. doi: 10.1111/cge.12754
- Hubacek M, Kripnerova T, Nemcikova M, et al (2016) Odontogenic keratocysts in the Basal Cell Nevus (Gorlin-Goltz) Syndrome associated with paresthesia of the lower jaw: Case report, retrospective analysis of a representative Czech cohort and recommendations for the early diagnosis. Neuro Endocrinol Lett 37:269–276.
- Bolar NA, Golzio C, Živná M, et al (2016) Heterozygous Loss-of-Function SEC61A1 Mutations Cause Autosomal-Dominant Tubulo-Interstitial and Glomerulocystic Kidney Disease with Anemia. Am J Hum Genet 99:174–87. doi: 10.1016/j.ajhg.2016.05.028
- Hartmannová H, Piherová L, Tauchmannová K, et al (2016) Acadian variant of Fanconi syndrome is caused by mitochondrial respiratory chain complex I deficiency due to a non-coding mutation in complex I assembly factor NDUFAF6. Hum Mol Genet. doi: 10.1093/hmg/ddw245
- Kousal B, Dudakova L, Gaillyova R, et al (2016) Phenotypic features of CRB1-associated early-onset severe retinal dystrophy and the different molecular approaches to identifying the disease-causing variants. Graefe’s Arch Clin Exp Ophthalmol 1–7. doi: 10.1007/s00417-016-3358-2
- Mizzi C, Dalabira E, Kumuthini J, et al (2016) A European Spectrum of Pharmacogenomic Biomarkers: Implications for Clinical Pharmacogenomics. PLoS One 11:e0162866. doi: 10.1371/journal.pone.0162866
- Neřoldová M, Stránecký V, Hodaňová K, et al (2016) Rare variants in known and novel candidate genes predisposing to statin-associated myopathy. Pharmacogenomics 17:1405–14. doi: 10.2217/pgs-2016-0071
- Stránecký V, Neřoldová M, Hodaňová K, et al (2016) Large copy-number variations in patients with statin-associated myopathy affecting statin myopathy-related loci. Physiol Res 1–19.
- Weidler S, Stopsack KH, Hammermann J, et al (2016) A product of immunoreactive trypsinogen and pancreatitis-associated protein as second-tier strategy in cystic fibrosis newborn screening. J Cyst Fibros. doi: 10.1016/j.jcf.2016.07.0
NCMG users
Charles University provides support to users of a wide range of specializations, both domestic and foreign. The NCMG infrastructure is also used in teaching and specialization education for students of bachelor's, master's, and doctoral programs, as well as for medical staff - physicians in the fields of medical genetics, pediatrics, gynecology and obstetrics, and specialists in laboratory methods - clinical genetics.
Domestic universities:
- 1.LF UK, Institute of Biochemistry and Experimental Oncology
- 2. LF UK, Institute of Biology and Medical Genetics
- 1.LF UK, KDDL, Laboratory of Eye Biology and Pathology
- Department of Pediatric Neurology2. LF UK and University Hospital Motol
- Department of Neurology, 2. LF UK and University Hospital Motol
- Department of Cardiology, 2. LF UK and University Hospital Motol
- Department of Urology, 2. LF UK and University Hospital Motol
- Department of Stomatology of Children and Adults, 2. LF UK and University Hospital Motol
- Department of Pediatrics, 2. LF UK and University Hospital Motol
- Department of Gynaecology and Obstetrics, 2. LF UK and University Hospital Motol
- Institute of Immunology, 2. LF UK and University Hospital Motol
- Department of Pathology and Molecular Medicine, 2. LF UK and University Hospital Motol
- Department of Surgery, 2. LF UK and University Hospital Motol
- Department of Dermatovenerology, 2. LF UK and University Hospital Motol
- Childhood Leukemia Investigation Prague, 2. LF UK
- Institute of medical microbiology, 2. LF UK
- Institute of Biology, Faculty of Medicine, Charles University in Pilsen
- University of West Bohemia in Pilsen - Faculty of Health Studies
Domestic public research organizations:
- Institute of Hematology and Blood Transfusion
- Institute of Biotechnology of the Czech Academy of Sciences
- Institute of Experimental Medicine of the Czech Academy of Sciences
- Institute of Molecular Genetics of the Czech Academy of Sciences
- IKEM, Prague, Department of Nephrology
- IKEM, Prague, Department of Cardiology
- IKEM, Prague, Department of Hepatogastroenterology
- Society of Medical Genetics and Genomics, Czech Medical Society of J. E. Purkyně
- Association of General Practitioners of the Czech Republic
Domestic health care providers:
- University Hospital Motol, Prague, Children's Cardiocentre
- University Hospital Motol, Prague, Laboratory of Molecular Genetics, Pediatric Clinic
- University Hospital Vinohrady, Prague
- University Hospital Motol, Department of Medical Molecular Genetics
- II. Department of Internal Medicine - Department of Cardiology and Angiology
- Psychiatric clinic of the General Teaching Hospital
- III. Internal Clinic of General Teaching Hospital - Department of Endocrinology and Metabolism
- Pediatric Nephrology, Department of Pediatric and Adolescent Medicine, General Teaching Hospital
- Department of Nephrology, General Teaching Hospital
- Department of Pediatric and Adolescent Medicine, General Teaching Hospital
- Mitochondrial Laboratory of General Teaching Hospital
- University Hospital Ostrava - Department of Medical Genetics
- University Hospital Hradec Králové - Department of Medical Genetics
Industrial enterprises:
- Pronatal.cz
- Repromeda.cz
- Gennet.cz
- VZP
- Biopharm
- Biogen
- GeneAge Technologies
- Multiplicom
- Vertex Pharmaceuticals
- Luminex
- Abbott Molecular
- IVF Center prof Zech Ltd.
- Bioptic laboratory s.r.o
Foreign universities, research institutions, and healthcare providers:
- Laboratory of Molecular and Medical Genetics, Faculty of Pure and Applied Sciences, University of Cyprus, Cyprus; Constantinos Deltas
- Clinical Genetics Department, National Research Center, Cairo, Egypt; Samia Temtamy
- Clinica Pediatrica, Monza, Italy; Angelo Selicorni
- Kinderklinik, Goettingen, Germany; Steinfeld
- Broad Institute, USA; Lucien Ronco
- Pediatric Nephrology Unit, University Hospital Reina Sofia, Cordoba, Spain; Montserrat Anton-Gamero
- Section on Nephrology, Wake Forest University School of Medicine, Winston-Salem, USA; A. Bleyer
- Nephrology Department, Hospital Fernando Fonseca, Portugal; Karin Soto
- Nephrology Department, Portsmouth Hospitals, Portmouth, United Kingdom; Dr. Venkat-Raman, Dr. Christine Gast
- Nephrology Unit, Guy's Hospital, London, United Kingdom; A. Marinaki
- Center for Human Genetics, Leuven, Belgium; Gert Matthijs
- Rare NCL Consortium, - a group of international laboratories
- University of Zagreb, prof. Sertic
- Children's Faculty Hospital in Košice - MUDr. Šaligová
- Slovak Academy of Sciences - Institute of Experimental Endocrinology Skopkova
- University of Uppsala
- University of Vita-Salute of San Raffaele
- Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry
- University of Szeged
- Center for Research and Technology Hellas - CERTH
- University of Oslo
Foreign industrial enterprises:
- Genzyme
Open Access
NCMG is a distributed multi-centered, nation-wide research infrastructure. The expertise and instrumentation available within NCMG can be used by the users from the Czech Republic and from abroad as well. It can be also used by the commercial sector. NCMG provides transparent information to their (potential) users on the access rules. NCMG through individual nodes (core facilities) support their users by providing an access to its instrumentation, analytical expertise, bioinformatic and statistical support, education and training and by providing an access to well defined cohorts of patients, control individuals and various clinical materials. Users have to adapt to the concrete access rules of the core facility they visit.
Users are obliged to acknowledge NCMG in any output (publications, patents, public presentations) that were created by using the NCMG instrumentation or expertise:
Since 2023:
"The National Center for Medical Genomics" or "The NCMG research infrastructure" (LM2023067)
Since 2020:
"The project "Modernization and instrumental upgrade of the National Center for Medical Genomics" (reg. No. CZ.02.1.01/0.0/0.0/18_046/0015515) is supported by the Operational Programme Research, Development and Education."
and
„We acknowledge the CF [name of the Core Facility] supported by the NCMG research infrastructure (LM2018132 funded by MEYS CR) for their support with obtaining scientific data presented in this paper.“
Before 2020:
"The authors would like to thank to The National Center for Medical Genomics (LM2015091) for providing allelic frequencies in ethnically matched population for comparison (project CZ.02.1.01/0.0/0.0/16_013/0001634)"
or
"We acknowledge the CF [name of the Core Facility] supported by the NCMG research infrastructure (LM2015091 funded by MEYS CR) for their support with obtaining scientific data presented in this paper."